Discovery of SoftSoap
SoftSoap Information
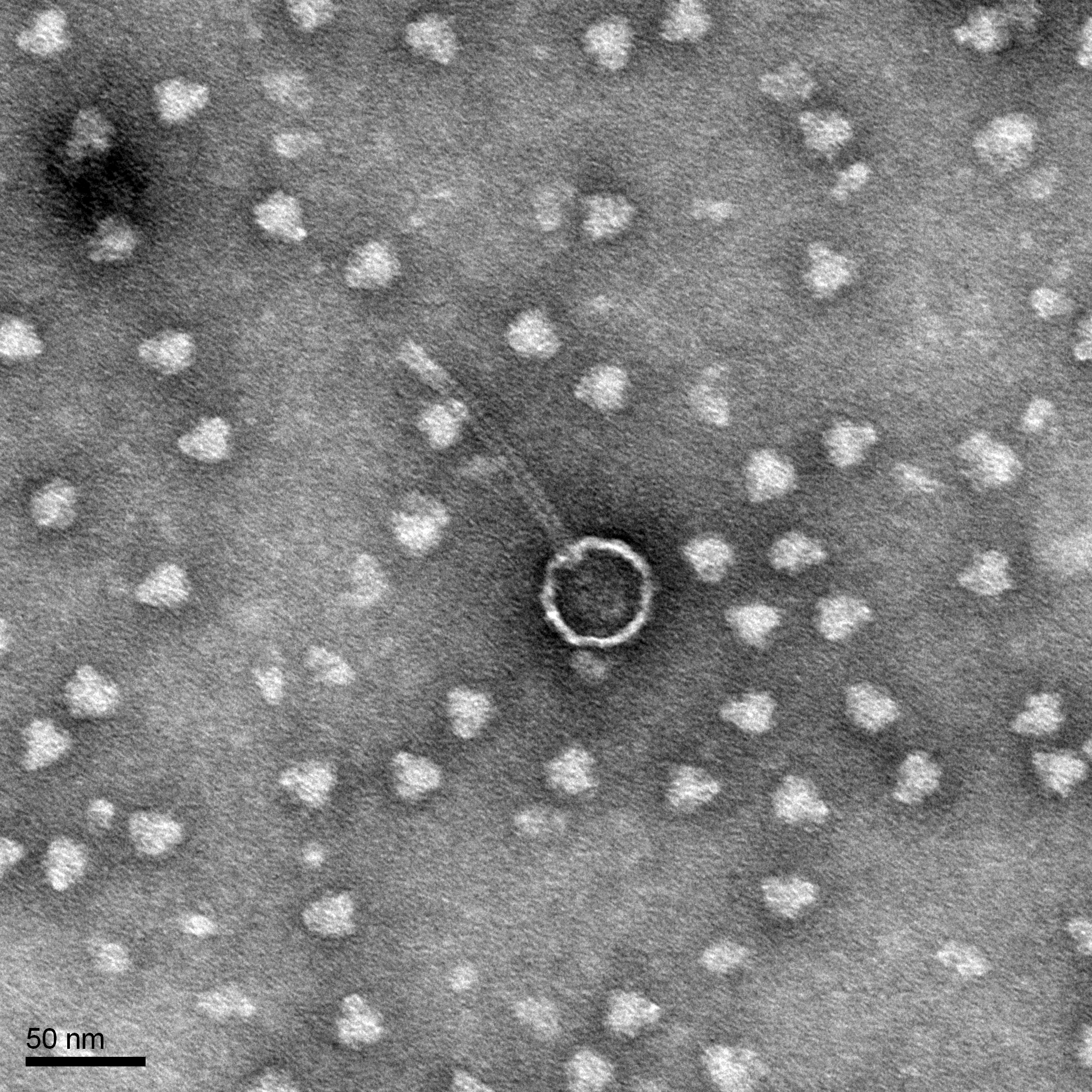
Morphology: Siphoviridae
Sample Collection
90
| Collector Name |
Hannah McNab | Kendall Brown | ||
| Sample No. | H1 | K1 | 3 | 4 |
| Date of Collection | 08.27.2024 | 08.27.2024 | ||
| Sample Type | soil | soil | soil | soil |
| General Location |
Under Oak Tree #3
|
beside a cow pond | ||
| Location Description | Underneath an oak tree. on a ranch in between 2 large roots | At the edge of a shallow water tank that has been used to water cows | ||
| GPS Coordinates | 30.97381 N, 97.60993 W | 32.40389 N, -98.14545 W | ||
| Sample Depth | 2 inches in the ground | surface level | ||
| Ambient Temperature | 21.6 celcius | 32 celcius |
Isolation/Purification
Sample H1
Date: 08/28/2024 Sample no. H1 Redo: No
Purpose: To isolate a bacteriophage from an environmental sample that will infect the host bacteria species.
Notes:
. Sample collection 08.28.2024
According to the techniques within the phage discovery guide section 5.2.
- First we collected soil samples into our 15 mL conical tubes, then we added enough fluid till the sample was submerged with about 3mL coverage with PYCa media.
- We inverted the tube a few times to ensure homogeneity within the sample. We then allowed the media-soil mixture to incubate for 1-2 hours in the shaking incubator.
- Then upon removal from the shaking incubator; the sample sat for ten minutes before continuing on to the next section
- Bacteriophage filtration 08.28.2024
- First we created an sterile environment by disinfecting our workspace with CiDeon, spraying then wiping it down with paper towels and then the same with 70% Ethanol Then we set up our bunsen burner and proceed to carefully work underneath the flame’s “umbrella”
- We attach the filter to the 5 mL syringe taking care to not contaminate the ends of either the filter nor the syringe. We remove the plunger from the 5 mL syringe and pour the supernatant off of the sample tube into the tube part of the syringe.
- Place the filter’s end on top of the microcentrifuge tube, reattach the plunger and push the liquid through the filter into the microcentrifuge.
- Cap the microcentrifuge and label
- Disposed of everything else, turn off the burner and reclean.
-
09.04.2024
Results: The H1 sample plate did have a couple pinpoint colonies so we proceeded to the phage dilution step of the phage discovery guide.
We picked a plaque from our plate. We sampled colony 1 to perform our dilutions according to the techniques detailed in the phage discovery guide 5.4 and 6.2.
Step one: we picked a plaque to isolate.
Observations: the colony that we selected was a small pinpoint colony round and smooth edged colony. It was very opaque when held up to the light. Please refer to the photo below. We used plaque 1.
The next steps would be to perform a serial dilution to ensure that we have only one pure phage plaque to work with. To do this we followed the phage discovery guide techniques 6.2.
Procedure: Serial dilutions
- We prepped with aseptic technique.
- Then acquired six microcentrifuge tubes we labeled them original and then 10-1 up to 10-5
- The we took 90 microliters of phage buffer in each of the tubes
- The using a p10 to spot the sample and then added it to the original microcentrifuge, then vortexed the sample
- Then carried 10 microliters to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.04.2024.
09.10.2024
Results; after 24 hours of incubation.
Observations: Observed pinpoint colonies on the H1 sample dilutions on the original plates and the 10-1plate, the K1 sample did not have any plaques at the 24 hour mark.
09.11.2024
Results: We observed the plaques on the H1 plates from the original all the way up to 10-4
. However we saw more clear plaques on the 10-2 plate so this is the plaque that we used.
After analyzing the results from round 1 of the serial dilutions we had an isolated phage on 10-2 and moved onto the second round of serial dilutions. Round 2 is to ensure the phage we extracted is completely isolated.
Procedure: Serial dilutions no. 2
- We prepped with aseptic technique.
- Then acquired six microcentrifuge tubes we labeled them original and then 10-1 up to 10-5
- The we took 90 microliters of phage buffer in each of the tubes
- The using a p10 to spot the sample and then added it to the original microcentrifuge, then vortexed the sample
- Then carried 10 microliters to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.04.2024.
Note: the dilutions of the phages sat for two hours at room temperature ( ) with the M.Foliorum bacterium before it was plated at 1220.
09.16.2024
09.16.2024
Results: we saw on our second round of dilutions that we needed to make one more set of dilutions in the hopes of growing a decent webbed plate to move one to the next step. So we made up dilutions round three.
Serial dilutions, no. 3
- We prepped with aseptic technique.
- Then acquired 5 microcentrifuge tubes we labeled them original and then 10-1 up to 10-4
- The we took 90 microliters of phage buffer in each of the tubes
- The using a p10 to spot the sample found in the 10-4 plate and then added it to the original microcentrifuge, then vortexed the sample
- Then carried 10 microliters to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.16.2024
09.18.2024
Results: We saw that on the third rounds of dilutions there had been an pipetting error between the 10-original and the 10-1 plate. Because of this we went back to look at the previous plates and looking at the second round of dilution’s original plate we found that it was sufficiently webbed and that we could move on to the next stage. According to the techniques described within the phage discovery program 6.3.
Phage Lysates. no.1
- Prepare the bench for aseptic work.
- Collect a webbed plate from the previous steps.
- Place 8 mL of Phage Buffer onto the webbed plate
- Gently swirl the plate ensuring that the entire plate is submerged.
- Allow the plate to sit for 12-14 hours in the fridge at 6 degrees celsius.
09.23.2024
Results: We came back to our plate, which had been incubating in the fridge at 6 degrees celsius. We then performed a Low Volume Isolate Spot Titer test titer, though instead of performing all dilutions on one single plate we used separate plates to better see the phages and identify the webbed plates. We used the techniques according to 6.2 serial dilutions and 5.2 for the plaque assay of the Phage Discovery program.
Completion of Lysate and the Full Plate Spot Titer!!
- After the incubation, allow the lysate to pool by gently tilting the plate.
- Using a 5 mL syringe aspirate the phage buffer out of the plate
- Attach the 0.22 uL filter and depress the syringe pouring the 15 mL conical tube.
- Repeat steps 7 and 8 till all of the phage buffer has been aspirated.
- Record the volume of how much lysate we have.
- Set up dilutions up to 10 -8 according to the techniques described in the phage discovery 6.2.
- Please refer to the section of the previous serial dilutions procedures.
(note: once lysate is removed it is good to store for up to one month)
Next Time: We hope to see several webbed plates by 09.25.2024.
09.25.2024
Results: The phages did not grow or swarm the plate as expected until the purification step was repeated using the same lysate. If this does not produce results we will remake dilutions and remake the lysate.
Procedure: ,Full plate spot titer 10^-3, off of new plaque off of 2nd round dilutions
- Set up dilutions up to 10 -8 according to the techniques described in the phage discovery 6.2.
- We prepped with aseptic technique.
- Then acquired four microcentrifuge tubes we labeled them original and then 10-1 up to 10-8
- The we took 90 microliters of PyCa media in each of the tubes
- Then we removed 10 ul of Lysate and placed into the original tube
- Then carried 10 microliters from the original to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.27.2024
We are left with 1.8 mL of Lysate.
09.27.2024
Results: The redo of the agar did not work, make up plates from round 2 serial dilutions picked a similar plaque.
Serial dilutions: 10-2
- We prepped with aseptic technique.
- Then acquired 4 microcentrifuge tubes we labeled them original and then 10-1 up to 10-2
- The we took 90 microliters of phage buffer in each of the tubes
- The using a p10 to spot the sample found in the 10-4 plate and then added it to the original microcentrifuge, then vortexed the sample
- Then carried 10 microliters to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
Set up three plates and let sit over the weekend.
09.30.2024
Results: The plaques set up last friday did not work. Picked new plaque from round 2 of serail dilutions (09.11.2024) and set up another round of dilutions.
Serial dilutions: 10-3
- We prepped with aseptic technique.
- Then acquired 4 microcentrifuge tubes we labeled them original and then 10-1 up to 10-3
- The we took 90 microliters of phage buffer in each of the tubes
- The using a p10 to spot the sample found in the 10-4 plate and then added it to the original microcentrifuge, then vortexed the sample
- Then carried 10 microliters to 10-1
- Repeat step 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 10.02.2024
10.02.2024
Results: The plates grew a webbed plate on the original plate.
Webbed plate Lysate:
Creating Phage Lysates. no.2
-
- Prepare the bench for aseptic work.
- Collect a webbed plate from the previous steps.
- Place 8 mL of Phage Buffer onto the webbed plate
- Gently swirl the plate ensuring that the entire plate is submerged.
- Allow the plate to sit for or 2-4 hours at room temperature 12-14 hours in the fridge at 6 degrees celsius.
- After the incubation, allow the lysate to pool by gently tilting the plate.
- Using a 5 mL syringe aspirate the phage buffer out of the plate
- Attach the 0.22 uL filter and depress the syringe pouring the 15 mL conical tube.
- Repeat steps 7 and 8 till all of the phage buffer has been aspirated.
- Record the volume of how much lysate we have.
Collected 1.8 ml of lysate
10.07.2024
Results: Full plate spot titer. Up to 10-3.
Procedure: Full Spot titer:
- Set up dilutions up to 10 -3 according to the techniques described in the phage discovery 6.2.
- We prepped with aseptic technique.
- Then acquired four microcentrifuge tubes we labeled them original and then 10-1 up to 10-3
- The we took 90 microliters of Phage buffer in each of the tubes
- Then we removed 10 ul of Lysate and placed into the original tube
- Then carried 10 microliters from the original to 10-1
- Repeat steps 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 10.09.2024
10.09.2024
Results: Weak full plate titer that only grew on the original plate. So we are now going to amplify our lysate.
Math:
Divided the original plate into quadrants,
Counted the phages in each quadrant for a totoal of 376 phage colonies.
376 x 10^3 x 1 (the dilution factor)
= 376,000 pfu/uL or 3.76 x 10 ^ 5 is our tube titer.
Amplification:
- We prepped with aseptic technique.
- Then acquired 7 microcentrifuge tubes we labeled them original and then 10+1 up to 10+6
- The we took 90 microliters of phage buffer in each of the tubes
- Then we added 10 uL of our Low titer lysate to the original, then 10 uL into each.
- Repeat step 4 till all samples have been amplified.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.11.2024
Plans: 10.11.2024, the plates were moved to the fridge to preserve them
10.14.2024
Results: The amplification was beautiful,and actually worked, we decided to use the 10 to the 7 plate because it was nice and webbed.
-
Procedure: Creating 6 identical plates: (Made up 7 of the 10 ^ 7)
- We prepped with aseptic technique.
- Then acquired 6 microcentrifuge tubes we labeled them 107 on all six.
- The we took 90 microliters of Phage buffer in each of the tubes
- Then we added 70 uL of our Low titer lysate to the all the tubes.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.21.2024
10.21.2024
Results: We forgot to grab the plates out of the incubator on Friday the 18, the plates did not turn out well this time either. They did not grow to a webbed plate. So we are going to amplify from 10^10 to 10^14. Based on the math that we should have been using.
Math:
So we need 5 x 10^9, phages on a plate. This means that if our lysate titer is at 3.76 x 10^5 we will need to amplify up to 10^13 and this should give us over 5×10^9 colonies.
Amplification!!-no.3
Procedure:
- We prepped with aseptic technique.
- Then acquired 5 microcentrifuge tubes we labeled them 10+10 up to 10+14
- The we took 90 microliters of phage buffer in each of the tubes
- Then we added 100 uL of our Low titer lysate, then increased by 10 uL of pure lysate into each.
- Repeat step 4 till all samples have been amplified.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.23.2024
Plans: We now hope for fully grown plates at 10.23.2024, from there we will create our 6 identical plates and be able to move on to the next step.
10.23.2024
Results: So, the 10^13 plate grew wonderfully, so did all of the other amplifications, however, after some input from the Professor, we will now make a new lysate that has a higher titer of phage inside of it.
Procedure: Six Identical plates.
- Clean the workspace aseptically
- Set up a bunsen burner
- Acquire 6 microcentrifuge tubes
- Aliquot 90 ul of Phage buffer to each, then add in 130 uL of the phage lysate
- Plate the 6 plates according to the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- By 10.25.2024 there should be 6 plates of reasonable webbing
Plans: For the next lab, flood 3 of the six plates, remove the supernatant and pour on to the other three plates. This will create a higher titer lysate that will be more functional, this will help the phages keep on ‘living’.
10/25/2024
Results: The plates all turned out webbed, so we will begin to make our third lysate. This third lysate will be completed 10.28.2024. This lysate is a more concentrated lysate than what we’ve had before. This lysate is from three webbed plates being flooded, then that lysate is then pooled into three other plates. This will create a much more concentrated lysate then what we were previously working with.
Procedure: Lysate
- Aseptically clean the workspace. And light the bunsen burner.
- Lay out three webbed plates and dispense 8 ml of Phage buffer onto each.
- Allow to sit for 3 hours and then remove the liquid off of the plate and onto the three other webbed plates that were made 10.23.2024
- Allow these to incubate in the refrigerator for the weekend, this lysate will be completed on 10/28/2024.
Plans: 10.28.2024, The lysate should be completed and a full plate spot titer should be run to calculate the titer of the new lysate. Technically this lysate would be our high titer lysate, assuming that the amount of phage is <5 x 10^9.
10.28.2024
Filtered out about 1 ml of the lysate, the lysate was very cloudy and extremely hard to filter. We successfully got about 1 ml of filtered lysate. We then set up dilution plates to figure out our titer. Will calculate the titer on 10.30.2024. The unfiltered high titer lysate is stored in the fridge.
Procedure: Full plate titers
- First we cleaned our area and created an aseptic environment to work in.
- Set up dilutions up to 10 -6 according to the techniques described in the phage discovery 6.2.
- Then acquired four microcentrifuge tubes we labeled them original and then 10-1 up to 10-6
- The we took 90 microliters of Phage Buffer media in each of the tubes
- Then we removed 10 ul of Lysate and placed into the original tube
- Then carried 10 microliters from the original to 10-1
- Repeat steps 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 10.09.2024
Plans: We hope to calculate our lysate’s titer on 10.30.2024
10.30.2024
Today we calculated our phage lysate and we also set up some new plates to create some more lysate for the future.
Math: On the 10-2 plate, 138 plaques were observed.
(138pfu/10uL) x (10^3) x (10^2)= 1.38 x 10^6
This is barely enough to begin the TEM microscopy.
Procedure: Six Identical plates.
- Clean the workspace aseptically
- Set up a bunsen burner
- Acquire 6 microcentrifuge tubes
- Aliquot 90 ul of Phage buffer to each, then add in 10 uL of the phage lysate
- Plate the 6 plates according to the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- By 11.01.2024 there should be 6 plates of reasonable webbing
- These we will flood with 10 ml of Phage Buffer instead of 8, because we require a little more and it soaks into the plates.
11.01.2024
Did the TEM microscopy according to the protocol in the Phage Discovery Guide 8.1b. Also I removed the plates from the incubator, parafilmed them and place in the fridge so that on Monday, we can create a more lysate of the same titer.
Procedure: TEM microscopy
- After asceptically cleaning the work space, and lighting the bunsen burner. While working under the flame; one ml of the high titer lysate was transferred into a steril microcentrifuge.
- The tubes where then centrifuged for one hour at top speed.
- Then the supernatant was removed, carefully not to disturb the button at the bottom.
- 100 uL of phage buffer was then added to the microcentrifuge. The button was resuspended and stored for 30 minutes at 4C (in the fridge)
- After the 30 minutes, fresh pair of gloves where put on.
- Work area under the hood was cleaned and lined with filter paper.
- The cover of a 5x5cm parafilm paper was removed, placed inside of a petri dish with Pleco tab on top.
- Then using EM forceps a fresh copper grid was removed, and placed shiny side up right next to the Pelco to where just an edge of the grid was touching the PELCO
- Then using a micropipetter,, 10uL of phage lysate onto the grid without touching the grid itself
- It was allowed to settle for 7 minutes
- Then using a small wedge of filter paper, the excess was wicked off.
- The grid was rinsed twice by first submerging it with 10 uL of steril water (allowing the water to sit for 2 minutes) and then wicking away the excess.
- After the water is wicked away, 10 ul of 1% Uranyl Acetate was added and allowed to sit for two minutes.
- The excess was wicked off, the surface looked like a rainbow oil slick by the time it was completely dry.
Plans: Monday we should make up the new lysate, so that we will have more to work with, we should also begin the protocols for DNA extraction starting at the 9.1 protocol.
Sample K1
Date: 09/04/2024 Redo: no Sample no. K1
Purpose: To isolate a bacteriophage from an environmental sample that will infect the host bacteria species.
Notes:
- Sample collection
According to the techniques within the phage discovery guide section 5.2.
- First we collected soil samples into our 15 mL conical tubes, then we added enough fluid till the sample was submerged with about 3mL coverage with PYCa media.
- We inverted the tube a few times to ensure homogeneity within the sample. We then allowed the media-soil mixture to incubate for 1-2 hours in the shaking incubator.
- Then upon removal from the shaking incubator; the sample sat for ten minutes before continuing on to the next section
- Bacteriophage filtration
- First we created an sterile environment by disinfecting our workspace with CiDeon, spraying then wiping it down with paper towels and then the same with 70% Ethanol Then we set up our bunsen burner and proceed to carefully work underneath the flame’s “umbrella”
- We attach the filter to the 5 mL syringe taking care to not contaminate the ends of either the filter nor the syringe. We remove the plunger from the 5 mL syringe and pour the supernatant off of the sample tube into the tube part of the syringe.
- Place the filter’s end on top of the microcentrifuge tube, reattach the plunger and push the liquid through the filter into the microcentrifuge.
- Cap the microcentrifuge and label
- Disposed of everything else, turn off the burner and reclean.
09.08.2024
- Plaque Assay 09.08.2024
- We prepared our work space using aseptic technique.
- Obtain an already prepped host bacteria from the TA
- Remove the K1 Sample’s microcentrifuge from the fridge.
- Aseptically dump the entirety of K1 sample into the bacteria’s tube
- Let sit for 10 minutes
- Using a sterile 5ml pipette remove 3 mL of the molten top agar and place into the tube of bacteria and potential viruses
- Give a quick mix
- Pour off onto the agar plate and tilt around in a circle so that the mixture completely covers the entirety of the plate
- Let the plates sit for 10-20 minutes or until the top agar has congealed.
- Allow to incubate for 24-48 hours
09.10.2024
Results; after 24 hours of incubation.
Observations: Observed pinpoint colonies on the H1 sample dilutions on the original plates and the 10-1plate, the K1 sample did not have any plaques at the 24 hour mark.
09.11.2024
Results: no plaques where observed on the plate, the plate was autoclaved and chalked up to a waste.
Amplification
10.09.2024
Results: Weak full plate titer that only grew on the original plate. So we are now going to amplify our lysate.
Math:
Divided the original plate into quadrants,
Counted the phages in each quadrant for a totoal of 376 phage colonies.
376 x 10^3 x 1 (the dilution factor)
= 376,000 pfu/uL or 3.76 x 10 ^ 5 is our tube titer.
Amplification:
- We prepped with aseptic technique.
- Then acquired 7 microcentrifuge tubes we labeled them original and then 10+1 up to 10+6
- The we took 90 microliters of PyCa media in each of the tubes
- Then we added 10 uL of our Low titer lysate to the original, then 10 uL into each.
- Repeat step 4 till all samples have been amplified.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.11.2024
Plans: 10.11.2024, the plates were moved to the fridge to preserve them
10.14.2024
Results: The amplification was beautiful,and actually worked, we decided to use the 10 to the 7 plate because it was nice and webbed.
-
Procedure: Creating 6 identical plates: (Made up 7 of the 10 ^ 7)
- We prepped with aseptic technique.
- Then acquired 6 microcentrifuge tubes we labeled them 107 on all six.
- The we took 90 microliters of PyCa media in each of the tubes
- Then we added 70 uL of our Low titer lysate to the all the tubes.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.21.2024
10.21.2024
Results: We forgot to grab the plates out of the incubator on Friday the 18, the plates did not turn out well this time either. They did not grow to a webbed plate. So we are going to amplify from 10^10 to 10^14. Based on the math that we should have been using.
Math:
So we need 5 x 10^9, phages on a plate. This means that if our lysate titer is at 3.76 x 10^5 we will need to amplify up to 10^13 and this should give us over 5×10^9 colonies.
Amplification!!-no.3
Procedure:
- We prepped with aseptic technique.
- Then acquired 5 microcentrifuge tubes we labeled them 10+10 up to 10+14
- The we took 90 microliters of PyCa media in each of the tubes
- Then we added 100 uL of our Low titer lysate, then increased by 10 uL of pure lysate into each.
- Repeat step 4 till all samples have been amplified.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- We now hope for fully grown plates at 10.23.2024
Plans: We now hope for fully grown plates at 10.23.2024, from there we will create our 6 identical plates and be able to move on to the next step.
10.23.2024
Results: So, the 10^13 plate grew wonderfully, so did all of the other amplifications, however, after some input from the Professor, we will now make a new lysate that has a higher titer of phage inside of it.
Procedure: Six Identical plates.
- Clean the workspace aseptically
- Set up a bunsen burner
- Acquire 6 microcentrifuge tubes
- Aliquot 90 ul of Phage buffer to each, then add in 130 uL of the phage lysate
- Plate the 6 plates according to the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- By 10.25.2024 there should be 6 plates of reasonable webbing
Plans: For the next lab, flood 3 of the six plates, remove the supernatant and pour on to the other three plates. This will create a higher titer lysate that will be more functional, this will help the phages keep on ‘living’.
10/25/2024
Results: The plates all turned out webbed, so we will begin to make our third lysate. This third lysate will be completed 10.28.2024. This lysate is a more concentrated lysate than what we’ve had before. This lysate is from three webbed plates being flooded, then that lysate is then pooled into three other plates. This will create a much more concentrated lysate then what we were previously working with.
Procedure: Lysate
- Aseptically clean the workspace. And light the bunsen burner.
- Lay out three webbed plates and dispense 8 ml of Phage buffer onto each.
- Allow to sit for 3 hours and then remove the liquid off of the plate and onto the three other webbed plates that were made 10.23.2024
- Allow these to incubate in the refrigerator for the weekend, this lysate will be completed on 10/28/2024.
Plans: 10.28.2024, The lysate should be completed and a full plate spot titer should be run to calculate the titer of the new lysate. Technically this lysate would be our high titer lysate, assuming that the amount of phage is <5 x 10^9.
10.28.2024
Filtered out about 1 ml of the lysate, the lysate was very cloudy and extremely hard to filter. We successfully got about 1 ml of filtered lysate. We then set up dilution plates to figure out our titer. Will calculate the titer on 10.30.2024. The unfiltered high titer lysate is stored in the fridge.
Procedure: Full plate titers
- First we cleaned our area and created an aseptic environment to work in.
- Set up dilutions up to 10 -6 according to the techniques described in the phage discovery 6.2.
- Then acquired four microcentrifuge tubes we labeled them original and then 10-1 up to 10-6
- The we took 90 microliters of Phage Buffer media in each of the tubes
- Then we removed 10 ul of Lysate and placed into the original tube
- Then carried 10 microliters from the original to 10-1
- Repeat steps 4 and 5 till all samples have been diluted.
- Then we referenced the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 10.09.2024
Plans: We hope to calculate our lysate’s titer on 10.30.2024
10.30.2024
Today we calculated our phage lysate and we also set up some new plates to create some more lysate for the future.
Math: On the 10-2 plate, 138 plaques were observed.
(138pfu/10uL) x (10^3) x (10^2)= 1.38 x 10^6
This is barely enough to begin the TEM microscopy.
Procedure: Six Identical plates.
- Clean the workspace aseptically
- Set up a bunsen burner
- Acquire 6 microcentrifuge tubes
- Aliquot 90 ul of Phage buffer to each, then add in 10 uL of the phage lysate
- Plate the 6 plates according to the phage guide 5.2 and plated the samples using the top agar that has been detailed on the Plaque Assay please reference on day 09.30.2024
- By 11.01.2024 there should be 6 plates of reasonable webbing
- These we will flood with 10 ml of Phage Buffer instead of 8, because we require a little more and it soaks into the plates.
11.01.2024
Did the TEM microscopy according to the protocol in the Phage Discovery Guide 8.1b. Also I removed the plates from the incubator, parafilmed them and place in the fridge so that on Monday, we can create a more lysate of the same titer.
Procedure: TEM microscopy
- After asceptically cleaning the work space, and lighting the bunsen burner. While working under the flame; one ml of the high titer lysate was transferred into a steril microcentrifuge.
- The tubes where then centrifuged for one hour at top speed.
- Then the supernatant was removed, carefully not to disturb the button at the bottom.
- 100 uL of phage buffer was then added to the microcentrifuge. The button was resuspended and stored for 30 minutes at 4C (in the fridge)
- After the 30 minutes, fresh pair of gloves where put on.
- Work area under the hood was cleaned and lined with filter paper.
- The cover of a 5x5cm parafilm paper was removed, placed inside of a petri dish with Pleco tab on top.
- Then using EM forceps a fresh copper grid was removed, and placed shiny side up right next to the Pelco to where just an edge of the grid was touching the PELCO
- Then using a micropipetter,, 10uL of phage lysate onto the grid without touching the grid itself
- It was allowed to settle for 7 minutes
- Then using a small wedge of filter paper, the excess was wicked off.
- The grid was rinsed twice by first submerging it with 10 uL of steril water (allowing the water to sit for 2 minutes) and then wicking away the excess.
- After the water is wicked away, 10 ul of 1% Uranyl Acetate was added and allowed to sit for two minutes.
- The excess was wicked off, the surface looked like a rainbow oil slick by the time it was completely dry.
Plans: Monday we should make up the new lysate, so that we will have more to work with, we should also begin the protocols for DNA extraction starting at the 9.1 protocol.
Characterization/Microscopy
TEM grid
11.01.2024
Did the TEM microscopy according to the protocol in the Phage Discovery Guide 8.1b. Also I removed the plates from the incubator, parafilmed them and place in the fridge so that on Monday, we can create a more lysate of the same titer.
Procedure: TEM microscopy
- After asceptically cleaning the work space, and lighting the bunsen burner. While working under the flame; one ml of the high titer lysate was transferred into a steril microcentrifuge.
- The tubes where then centrifuged for one hour at top speed.
- Then the supernatant was removed, carefully not to disturb the button at the bottom.
- 100 uL of phage buffer was then added to the microcentrifuge. The button was resuspended and stored for 30 minutes at 4C (in the fridge)
- After the 30 minutes, fresh pair of gloves where put on.
- Work area under the hood was cleaned and lined with filter paper.
- The cover of a 5x5cm parafilm paper was removed, placed inside of a petri dish with Pleco tab on top.
- Then using EM forceps a fresh copper grid was removed, and placed shiny side up right next to the Pelco to where just an edge of the grid was touching the PELCO
- Then using a micropipetter,, 10uL of phage lysate onto the grid without touching the grid itself
- It was allowed to settle for 7 minutes
- Then using a small wedge of filter paper, the excess was wicked off.
- The grid was rinsed twice by first submerging it with 10 uL of steril water (allowing the water to sit for 2 minutes) and then wicking away the excess.
- After the water is wicked away, 10 ul of 1% Uranyl Acetate was added and allowed to sit for two minutes.
- The excess was wicked off, the surface looked like a rainbow oil slick by the time it was completely dry.
Results:
Characterization
08.30.2024
Purpose: To differentiate between the other phages on the plate.
Notes: the colony that we selected was a small pinpoint colony round and smooth edged colony. It was very opaque when held up to the light.
DNA Extraction
of
Precipitating DNA
11/11/24 DNA Extraction
- After gently mixing our high volume lyaste we placed 5mL the HVL into 15mL conical tubes and added 20 uL of nuclease.
- then we gently inverted the tube and incubated for 10 minuets at 37 degrees Celsius.
- The we took 1mL of lyaste and placed it into 4 microcentrifuge tubes
- we added 20 uL of ZnCl2 into each microcentrifuge tubes and mixed it gently by inverting it a 3 times.
- After inverting each tube we place them into a centrifuge at 10,000 rpm for 1 minute this pelleted the phage
- then removed all the supernatent by aspirating making sure not to mess with the DNA pallete
- we then resuspended the pellets in 500 uL of TES buffer and incubated at 60 degrees Celsius for 15 minutes
- After incubating we then added 1 uL of Proteinase K and mixed gently by inverting. It was placed in the incubator at 37 degrees Celsius for 10 minutes
- next we added 60 uL of Potassium acetate to all the tubes mixed well by inverting a few times and left on ice for 15 minutes
- once it was done resting in ice we added it to a 4 degree Celsius centrifuge at 12,000 rpm for 1 minute. After this we kept the supernatant and placed it into new microcentrifuge tubes
- next we added 500 uL of isopropanol to all the tubes and let rest in a ice bucket until our next lan on 11/13/2024
11/13/2024 DNA Extraction part 2
- We removed all the tubes from the ice bucket and placed it in the centrifuge at max speed for 10 minutes. 3 of the 4 tubes formed a pellet but it was ok if no pellet appeared.
- Then we added 250 uL of 70% ethanol to the tube, this was spun again at top speed and then the supernatants where removes.
- The tubes where left open on a paper towel to dry for 40 minutes.
- Then using 50ul of nuclease-free water the first DNA pellet was resuspended. Then removing the same nuclease free water from that tube, the water was pitpetted into the next one and so on down the line. (there are only 4)
- The DNA was taken to the Nano drop for the readings.
Results: The concentration of DNA was 150.5 ng/uL
T A260/A280 is 1.75
the A260/A230 is 0.92
11/15/2024
Made up more DNA extraction. And Added Enzymes to the new DNA to run on Tuesday. Created more lysate just in case
Procedure:
- After gently mixing our high volume lyaste we placed 5mL the HVL into 15mL conical tubes and added 20 uL of nuclease.
- then we gently inverted the tube and incubated for 10 minuets at 37 degrees Celsius.
- The we took 1mL of lyaste and placed it into 4 microcentrifuge tubes
- we added 20 uL of ZnCl2 into each microcentrifuge tubes and mixed it gently by inverting it a 3 times.
- After inverting each tube we place them into a centrifuge at 10,000 rpm for 1 minute this pelleted the phage
- then removed all the supernatent by aspirating making sure not to mess with the DNA pallete
- we then resuspended the pellets in 500 uL of TES buffer and incubated at 60 degrees Celsius for 15 minutes
- After incubating we then added 1 uL of Proteinase K and mixed gently by inverting. It was placed in the incubator at 37 degrees Celsius for 10 minutes
- next we added 60 uL of Potassium acetate to all the tubes mixed well by inverting a few times and left on ice for 15 minutes
- once it was done resting in ice we added it to a 4 degree Celsius centrifuge at 12,000 rpm for 1 minute. After this we kept the supernatant and placed it into new microcentrifuge tubes
- next we added 500 uL of isopropanol to all the tubes and let rest in a ice bucket until our next lan on 11/25/2024
11/18/2024
DNA reprecipitation
Extraction Part 2: removed the tubes from the freezer where they were frozen at negative 20 for the weekend. Set up the restriction enzymes to be run on Tuesday the 26.
DNA Extraction part 2
- We removed all the tubes from the ice bucket and placed it in the centrifuge at max speed for 10 minutes. 3 of the 4 tubes formed a pellet but it was ok if no pellet appeared.
- Then we added 250 uL of 70% ethanol to the tube, this was spun again at top speed and then the supernatants where removes.
- The tubes where left open on a paper towel to dry for 40 minutes.
- Then using 50ul of nuclease-free water the first DNA pellet was resuspended. Then removing the same nuclease free water from that tube, the water was pitpetted into the next one and so on down the line. (there are only 4)
- The DNA was taken to the Nano drop for the readings.
Nano readings:
DNA concentration was 746.0 ng/uL
The A260/A280 is 1.86
The A260/A230 is 1.86
This is good enough to run a gel on.
Setting up restriction enzymes and running the gel
1. After setting up a clean work space, we acquired 7 microcentrifuge tubes. One for the ladder, one for uncut and 5 for the restriction enzymes.
2. After we left everything to thaw to room temperature, We set the DNA to incubate at 60 degrees Celsius for ten minutes.
3. After labelling all the tubes we removed 3ul of the DNA and placed into 6 of the tubes labels Uncut, Hae3, Mse1, Nsp1, Sac2, and Sal1. Then to the Enzyme tubes we added 1ul of the corresponding enzyme and 2 ul their associated buffer and 14 ul of water. For the Uncut we only added 17 of water to the tube.
4. The mixtures where placed in the freezer for storage at -20 Celsius.
11/19/2024
Procedure:
1. The gel running day! We ran the Gel that was made by pouring in 30ml of 1TEA Buffer and 0. 24 g of Argarose. These where mixed in a erlenmeyer flask and microwaved for about 30 seconds and swirled till clear.
2. The gel was then poured into the mold and allow to set.
3. Then the DNA tubes and enzymes were allowed to thaw and 5 ul of gel loading dye was added to each tube. Each tube was vortexed and centrifuged to mix then 20 ul of sample was added into seven different wells.
4. The electrophoresis was run for 1 hour and then the gel was removed and then stained with 0.1% Ethidium Bromide. Then rinsed with DI water twice.
5. The gel was read under a UV light,
Results: Faint bands where seen on the uncut and the Hae3 rows. The gel is going to be rerun for perhaps a clearer image.
11/20/2024
Procedure:
1. The gel running day! We ran the Gel that was made by pouring in 30ml of 1TEA Buffer and 0. 24 g of Argarose. These where mixed in a erlenmeyer flask and microwaved for about 30 seconds and swirled till clear.
2. The gel was then poured into the mold and allow to set.
3. Then the DNA tubes and enzymes were allowed to thaw and 5 ul of gel loading dye was added to each tube. Each tube was vortexed and centrifuged to mix then 20 ul of sample was added into seven different wells.
4. The electrophoresis was run for 1 hour and then the gel was removed and then stained with 0.1% Ethidium Bromide. Then rinsed with DI water twice.
5. The gel was read under a UV light,
Results; The bands were not visible. we will try one more time.
11/22/2024
Procedure:
1. The gel running day! We ran the Gel that was made by pouring in 30ml of 1TEA Buffer and 0. 24 g of Argarose. These where mixed in a erlenmeyer flask and microwaved for about 30 seconds and swirled till clear.
2. The gel was then poured into the mold and allow to set.
3. Then the DNA tubes and enzymes were allowed to thaw and 5 ul of gel loading dye was added to each tube. Each tube was vortexed and centrifuged to mix then 20 ul of sample was added into seven different wells.
4. The electrophoresis was run for 1 hour and then the gel was removed and then stained with 0.1% Ethidium Bromide. Then rinsed with DI water twice.
5. The gel was read under a UV light,
Results: This gel also did not show the DNA, so we are using the original run and sending it out.