Discovery of Ganandorf
Ganandorf Information
Morphology: Siphoviridae
Sample Collection
| Collector Name |
Elijah Rodriguez | Elijah Rodriguez | Katelyn Kaase | Elijah Rodriguez | Katelyn Kaase | Elijah Rodriguez |
| Sample No. | 1 | 2 | 3 | 4 | 5 | 6 |
| Date of Collection | 08/29/2023 | 09/05/2023 | 9/05/2023 | 9/12/2023 | 09/18/2023 | 9/19/2023 |
| Sample Type | Soil | Soil | Soil | Soil | Soil | Soil |
| General Location | Flower Pot Soil | Backyard of collector home | Back of Subway’s parking lot | The front yard of collector home | Nursing Building side of the Science Building at TSU | Side yard of the collector home |
| Location Description | The sample was collected in a flower pot, which was located in the front of the collector’s house. | The sample was collected behind the in-ground pool in the backyard of the collector’s house. | The sample was collected in a dirt pile behind the back of the Subway’s parking lot. | The sample was collected in the soil in front of the collector’s house. | The sample was located on an ant hill about 10 feet away from the side door of the Science building that faces the Nursing building. | The sample was located in an anthill in the sideyard of the collector’s home. |
| GPS Coordinates | 32.25634° N, 98.21932° W | 32.25634° N, 98.21932° W | 32.214408 N, 98.213473 W | 32.25634° N, 98.21932° W | 32.21678°N, 98.22055°W | 32.25634° N, 98.21932° W |
| Sample Depth | 3 cm | 2 cm | Surface level | 5 cm | Surface to 2 cm deep | Surface to 2 cm deep |
| Ambient Temperature | 20°C | 30.5556°C | 35 C | 20°C | 35°C | 23.9°C |
Isolation/Purification
Title: Direct Isolation/Purification of Environmental Sample
Date: 08/30/23
Redo: No
Sample: #1
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin Direct Isolation.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 15 ml conical tube
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol.
- Approximately 1/3 of a 15 ml conical tube was filled with the soil sample.
- 15ml of a sterile 50 ml conical tube was filled with the soil sample.
- The conical tube was capped and manually mixed thoroughly.
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for 1 hour.
- Once the hour had passed, the tube was taken out of the incubator and left undisturbed for approximately 5 minutes to allow the sample inside to settle.
- Once settled, a package of syringe filter (0.22μm) was opened and the filter was left in the packaging for later use.
- A Bunsen burner was setup and lit to create a radial sterile field to prevent contamination to the sample and the filter.
- Using the syringe, approximately 2 ml of liquid was collected from the top of of the flooded conical tube containing the sample while minimizing collection of any debris to prevent clogging during the filter filtration.
- The syringe was then attached to the top of the filter, making sure it was screwed in place, and the filter was removed from the packaging to prevent contamination.
- The syringe plunger was depressed, and .5 ml of filtrate was collected into a micro-centrifuge tube. (note: to prevent contamination, the liquid was filtered slowly as to not break the filter or force liquid through the filter.)
- The tube was then capped immediately to prevent contamination and the syringe and filter were discarded.
Results:
After capping of the labeled micro-centrifuge tube, the solution is described as a clear color.
Conclusions and Next Steps:
Once the labeled micro-centrifuge tube was capped, the tube was placed on the bench and the supplies were assembled to begin the Plaque Assay procedure.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 08/30/23
Redo: No
Sample: #1
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- Prior to procedure, the supplies used for Purification of Environmental Sample by Plaque Assay were assembled.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
Procedure:
- Once the tube was capped following direct isolation, a test tube containing 250μl host bacterial cultures was collected as well as an agar plate.
- Once the tube was collected, using a micropipettor, the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria and was gently mixed.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for use.
- A bottle of top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube. (Note: make sure all the equipment is placed under the sterile field created by the bunsen burner to keep the equipment from becoming contaminated).
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate and the pipette is discarded. (Important: Try to avoid making or sucking up bubbles, since they can look like plaques on the agar plates).
- The agar plate is gently and quickly tilted in multiple directions to evenly coat the agar plate in the top agar mixture.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 24-48 hours at a temperature of 29˚C .
Results:
– After 24 hours of incubation, the plate began to show a small amount of contamination near the center of the plate and no plaques were formed.
– After 48 hours of incubation, the plate showed signs of contamination with no formation of plaques and the procedure was deemed unsuccessful, where the plate was then discarded.
Direct Isolation and Plaque Assay of Sample 1
Conclusions and Next Steps:
Direct Isolation and Purification of Environmental sample by Plaque Assay of sample #1 showed unsatisfactory results with no formation of plaques on the plate.
Next steps include the collection of a new soil sample and beginning the Enriched Isolation/Purification of Environmental Sample procedure.
Title: Enriched Isolation/Purification of Environmental Sample
Date: 09/06/23
Redo: No
Sample: #3
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- All steps of the procedure were worked under the radical sterile environment that the Bunsen burner created to ensure contamination would not occur.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 50 ml conical tube
- .5 mL host bacteria (M. foliorum)
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace CiDecon and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol. After allowing the disinfectant dried throughly, a Bunsen Burner was lit to create a radical sterile environment.
- 15 mL of the environmental sample was transferred to a 50 mL sterile conical tube.
- Approximately 20mL of liquid media is added to the tube (to fill it to the 35 mL mark)
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for an hour.
- Once the hour had passed, the tube was transferred to a centrifuge, balanced, and pelleted at 2,000 g for 10 minutes.
- After being cetrifuged, a vaccuum filtration unit was used to extract the liquid sample and filter out any debris.
- 0.5 mL of host bacteria M. foliorum was added to the 50 mL conical tube containing our filtered liquid sample.
- We made sure our conical tube was properly aerated by twisting the cap one quarter of a turn before securely taping down the cap to prevent it from falling off.
- The tube was then placed in a shaking incubator for 5 days.
- After the 5 days, we returned to the lab (9/11/2023), prepped our workspace by creating an aseptic environment, and grabbed our sample from the incubator.
- Using a 5 mL serological pipette, we transferred 1.4 mL of our enriched culture into a microcentrifuge tube. This step was repeated so that we had two 1.4 mL samples in separate microcentrifuge tubes.
- We then spun the two microcentrifuge tubes in a microcentrifuge for 1 minute.
- Our samples were not clear enough for our liking, so we filtered our samples through a 0.22 μm syringe filter. 1 mL of each sample was inserted into the syringe barrel before being filtered into a clean microcentrifuge tube.
- After immediately capping and labeling our microcentrifuge tube containing our filtered samples, we set the tube aside to begin the plaque assay.
Results:
After capping of the labeled micro-centrifuge tube, the solution is described as a clear color. We will not know for certain well our samples produce plaques until we return to lab.
Conclusions and Next Steps:
Once the labeled micro-centrifuge tube was capped, the tube was placed on the bench and the supplies were assembled to begin the Plaque Assay procedure.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 09/11/23
Redo: No
Sample: #3
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- All steps of the procedure were worked under the radical sterile environment that the Bunsen burner created to ensure contamination would not occur.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
- Micropipette
Procedure:
- Once the tube was capped following enriched isolation, a test tube containing 10 μl host bacterial cultures was collected as well as an agar plate.
- Using a micropipettor, the entirety of the sample in the microcentrifuge tube (2 mL) was transferred into the tube containing the host bacteria M. foliorum and was gently mixed by tapping the tube.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for future use.
- A bottle of molten top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube, working very hard to work quickly.
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate, working hard to avoid making or sucking up bubbles, as they can be confused for plaques on the agar plates. We then immediately discarded the pipette.
- The agar plate is gently and quickly tilted in multiple directions to give the agar plate an even and level coat of the molten top agar.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 48 hours at a temperature of (29°C).
Results:
– After 24 hours of incubation, the sample showed unsatisfactory results with no signs of plaques on the agar plate. A bubble had formed during the top agar step, which is shown in the center of the plate on the picture. Picture is shown below:
24 Hours plaque assay for Kates 1st enrichment sample
– After 48 hours of incubation, the sample showed unsatisfactory results with no signs of plaques on the plate. Picture is shown below:
Plaque Assay 72 hour for Kates Enrichment sample as of September 13
Conclusions and Next Steps:
– Following unsuccessful results of the enrichment procedure for sample #3, we will conduct another enriched isolation of sample #4 in an attempt to discover a bacteriophage. Additionally, a direct isolation procedure of sample #2 will be conducted to attempt discovery.
Title: Direct Isolation/Purification of Environmental Sample
Date: 09/11/23
Redo: Yes
Sample: #2
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin Direct Isolation.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 15 ml conical tube
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol.
- Approximately 1/3 of a 15 ml conical tube was filled with the soil sample.
- 15ml of a sterile 50 ml conical tube was filled with the soil sample.
- The conical tube was capped and manually mixed thoroughly.
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for 1 hour.
- Once the hour had passed, the tube was taken out of the incubator and left undisturbed for approximately 5 minutes to allow the sample inside to settle.
- Once settled, a package of syringe filter (0.22μm) was opened and the filter was left in the packaging for later use.
- A Bunsen burner was setup and lit to create a radial sterile field to prevent contamination to the sample and the filter.
- Using the syringe, approximately 2 ml of liquid was collected from the top of of the flooded conical tube containing the sample while minimizing collection of any debris to prevent clogging during the filter filtration.
- The syringe was then attached to the top of the filter, making sure it was screwed in place, and the filter was removed from the packaging to prevent contamination.
- The syringe plunger was depressed, and .5 ml of filtrate was collected into a micro-centrifuge tube. (note: to prevent contamination, the liquid was filtered slowly as to not break the filter or force liquid through the filter.)
- The tube was then capped immediately to prevent contamination and the syringe and filter were discarded.
Results:
– After capping of the labeled micro-centrifuge tube, the solution is described as a clear color.
Conclusions and Next Steps:
– Once the labeled micro-centrifuge tube was capped, the tube was placed on the bench and the supplies were assembled to begin the Plaque Assay procedure.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 09/11/23
Redo: Yes
Sample: #2
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- Prior to procedure, the supplies used for Purification of Environmental Sample by Plaque Assay were assembled.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
Procedure:
- Once the tube was capped following direct isolation, a test tube containing 250μl host bacterial cultures was collected as well as an agar plate.
- Once the tube was collected, using a micropipettor, the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria and was gently mixed.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for use.
- A bottle of top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube. (Note: make sure all the equipment is placed under the sterile field created by the bunsen burner to keep the equipment from becoming contaminated).
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate and the pipette is discarded. (Important: Try to avoid making or sucking up bubbles, since they can look like plaques on the agar plates).
- The agar plate is gently and quickly tilted in multiple directions to evenly coat the agar plate in the top agar mixture.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 24-48 hours at a temperature of 29˚C .
Results:
– Following the 24 hour period, the sample was collected and placed in front of a light to locate signs of plaques, where none were discovered. In the center and top of the plate, bubbles were formed while performing the top agar step. Picture is shown below:
Plaque Assay 24 hour for Elijah’s direct isolation september 13
– Following the 48 hour period, the sample was once again collected and placed in front of a light to locate signs of plaques, where none were discovered, deeming the procedure unsuccessful. Picture is shown below:
Plaque Assay 72 hour for Elijah direct isolation as of september 13
Conclusion and Next Steps:
– After the unsuccessful experiment, the sample was discarded and we began the preparation for the second attempt of the Enriched Isolation procedure using sample #4.
Title: Enriched Isolation/Purification of Environmental Sample
Date: 09/13/23
Redo: Yes
Sample: #4
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- All steps of the procedure were worked under the radical sterile environment that the Bunsen burner created to ensure contamination would not occur.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 50 ml conical tube
- .5 mL host bacteria (M. foliorum)
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace CiDecon and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol. After allowing the disinfectant dried throughly, a Bunsen Burner was lit to create a radical sterile environment.
- 15 mL of the environmental sample was transferred to a 50 mL sterile conical tube.
- Approximately 20mL of liquid media is added to the tube (to fill it to the 35 mL mark)
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for an hour.
- Once the hour had passed, the tube was transferred to a centrifuge, balanced, and pelleted at 2,000 g for 10 minutes.
- After being cetrifuged, a vaccuum filtration unit was used to extract the liquid sample and filter out any debris.
- 0.5 mL of host bacteria M. foliorum was added to the 50 mL conical tube containing our filtered liquid sample.
- We made sure our conical tube was properly aerated by twisting the cap one quarter of a turn before securely taping down the cap to prevent it from falling off.
- The tube was then placed in a shaking incubator for 2 days.
- After the 2 days, we returned to the lab (9/15/2023), prepped our workspace by creating an aseptic environment, and grabbed our sample from the incubator.
- Using a 5 mL serological pipette, we transferred 1.4 mL of our enriched culture into a microcentrifuge tube. This step was repeated so that we had two 1.4 mL samples in separate microcentrifuge tubes.
- We then spun the two microcentrifuge tubes in a microcentrifuge for 1 minute.
- Our samples were not clear enough for our liking, so we filtered our samples through a 0.22 μm syringe filter. 1 mL of each sample was inserted into the syringe barrel before being filtered into a clean microcentrifuge tube.
- After immediately capping and labeling our microcentrifuge tube containing our filtered samples, we set the tube aside to begin the plaque assay.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 09/15/23
Redo: Yes
Sample: #4
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- Prior to procedure, the supplies used for Purification of Environmental Sample by Plaque Assay were assembled.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
Procedure:
- Once the tube was capped following direct isolation, a test tube containing 250μl host bacterial cultures was collected as well as an agar plate.
- Once the tube was collected, using a micropipettor, the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria and was gently mixed.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for use.
- A bottle of top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube. (Note: make sure all the equipment is placed under the sterile field created by the bunsen burner to keep the equipment from becoming contaminated).
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate and the pipette is discarded. (Important: Try to avoid making or sucking up bubbles, since they can look like plaques on the agar plates).
- The agar plate is gently and quickly tilted in multiple directions to evenly coat the agar plate in the top agar mixture.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 24-48 hours at a temperature of 29˚C .
Results:
– The results of our enriched isolation procedure were deemed unsatisfactory following a 48 hour period as no signs of plaque development were prevalant as shown in photo below. We will repeat the procedure through direct isolation. The following picture will show a large scratch as the sample was tampered with prior to viewing due to miscommunication.
48 hour Plaque Assay for Sample 4 (1)
Conclusions and Next Steps: We will redo an attempt at Direct Isolation/Purification of another environmental sample to attempt to locate signs of bacteriophages.
Title: Direct Isolation/Purification of Environmental Sample
Date: 09/18/23
Redo: Yes
Sample: #5
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin Direct Isolation.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 15 ml conical tube
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol.
- Approximately 1/2 of a 15 ml conical tube was filled with the soil sample.
- Liquid media was added to the 15 mL conical tube until the soil sample is submerged under approximately 3 mL.
- The conical tube was capped and manually shaken.
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for 1 hour.
- Once the hour had passed, the tube was taken out of the incubator and left undisturbed for approximately 5 minutes to allow the sample inside to settle.
- A Bunsen burner was lit to create a radical sterile environment.
- Once settled, a package of syringe filter (0.22μm) was opened and the filter was left in the packaging for later use.
- Using the syringe, approximately 2 ml of liquid was collected from the top of of the flooded conical tube containing the sample while minimizing collection of any debris to prevent clogging during the filter filtration.
- The syringe was then attached to the top of the filter, making sure it was screwed in place, and the filter was removed from the packaging to prevent contamination.
- The syringe plunger was depressed, and .5 ml of filtrate was collected into a micro-centrifuge tube. (note: to prevent contamination, the liquid was filtered slowly as to not break the filter or force liquid through the filter.)
- The tube was then capped immediately to prevent contamination and the syringe and filter were discarded.
Results:
– After capping of the labeled micro-centrifuge tube, the solution is described as a clear color.
Conclusions and Next Steps:
– Once the labeled micro-centrifuge tube was capped, the tube was placed on the bench and the supplies were assembled to begin the Plaque Assay procedure.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 09/18/23
Redo: Yes
Sample: #5
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- Prior to procedure, the supplies used for Purification of Environmental Sample by Plaque Assay were assembled.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
Procedure:
- Once the tube was capped following direct isolation, a test tube containing 250μl host bacterial cultures was collected as well as an agar plate.
- Once the tube was collected, using a micropipettor, the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria and was gently mixed.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for use.
- A bottle of top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube. (Note: make sure all the equipment is placed under the sterile field created by the bunsen burner to keep the equipment from becoming contaminated).
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate and the pipette is discarded. (Important: Try to avoid making or sucking up bubbles, since they can look like plaques on the agar plates).
- The agar plate is gently and quickly tilted in multiple directions to evenly coat the agar plate in the top agar mixture.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 24-48 hours at a temperature of 29˚C .
Results:
– Following 24 hours of incubation, the sample showed no signs of plaques as shown in the picture below:
Plaque Assay 24 hour for direct Isolation of sample 5
– The sample was placed back in the incubator for another 24 hours to allow time for plaques to form.
– After 48 hours of incubation, the sample was taken out and examined, which showed no signs of plaques formed on the agar plate as shown in the picture below; deeming the experiment unsucessful.
Plaque Assay 48 hour results for sample 5
Conclusions and Next Steps:
– Following the unsucessful experiment, the agar plate was discarded and preparations were made for another attempt at Direct Isolation/Purification of another Environmental sample to attempt to locate signs of bacteriophages.
Title: Direct Isolation/Purification of Environmental Sample
Date: 09/20/23
Redo: Yes
Sample: #6
Purpose: To extract bacteriophages from microbes and matter located in the soil sample.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin Direct Isolation.
Supplies:
-
- Environmental sample
- Liquid media (5 ml/sample)
- Sterile 3 ml or 5 ml syringe
- 0.22 μm syringe filter
- 5 ml serological pipettes
- Microcentrifuge tubes
- 15 ml conical tube
Procedure:
-
- To create an aseptic environment, we put on gloves and sprayed our workspace and wiped it dry before allowing it to evaporate. These steps were repeated with 70% ethanol.
- Approximately 1/2 of a 15 ml conical tube was filled with the soil sample.
- Liquid media was added to the 15 mL conical tube until the soil sample is submerged under approximately 3 mL.
- The conical tube was capped and manually shaken.
- The conical tube containing the mixture was placed in a shaking incubator, which was shaken vigorously at 250 rpm for 1 hour.
- Once the hour had passed, the tube was taken out of the incubator and left undisturbed for approximately 5 minutes to allow the sample inside to settle.
- A Bunsen burner was lit to create a radical sterile environment.
- Once settled, a package of syringe filter (0.22μm) was opened and the filter was left in the packaging for later use.
- Using the syringe, approximately 2 ml of liquid was collected from the top of of the flooded conical tube containing the sample while minimizing collection of any debris to prevent clogging during the filter filtration.
- The syringe was then attached to the top of the filter, making sure it was screwed in place, and the filter was removed from the packaging to prevent contamination.
- The syringe plunger was depressed, and .5 ml of filtrate was collected into a micro-centrifuge tube. (note: to prevent contamination, the liquid was filtered slowly as to not break the filter or force liquid through the filter.)
- The tube was then capped immediately to prevent contamination and the syringe and filter were discarded.
Results:
– After capping of the labeled micro-centrifuge tube, the solution is described as a clear color.
Conclusions and Next Steps:
– Once the labeled micro-centrifuge tube was capped, the tube was placed on the bench and the supplies were assembled to begin the Plaque Assay procedure.
Title: Isolation/Purification of Environmental Sample by Plaque Assay
Date: 09/20/23
Redo: Yes
Sample: #6
Purpose: To detect the presence of bacteriophages on bacterial lawns.
Notes:
- Prior to procedure, the supplies used for Purification of Environmental Sample by Plaque Assay were assembled.
Supplies:
-
- Phage samples for isolation, purification, or titering
- Host bacteria (250 μl/plate)
- Agar plates
- Phage buffer
- Top agar, molten (between 55 – 60 ˚C)
- Microcentrifuge tubes
- 5 ml serological pipettes
Procedure:
- Once the tube was capped following direct isolation, a test tube containing 250μl host bacterial cultures was collected as well as an agar plate.
- Once the tube was collected, using a micropipettor, the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria and was gently mixed.
- Once the tube was mixed, the tube was left undisturbed for around 5 minutes to allow for attachment.
- The agar plate is then labeled accordingly and set on the table for use.
- A bottle of top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacteria culture tube. (Note: make sure all the equipment is placed under the sterile field created by the bunsen burner to keep the equipment from becoming contaminated).
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate and the pipette is discarded. (Important: Try to avoid making or sucking up bubbles, since they can look like plaques on the agar plates).
- The agar plate is gently and quickly tilted in multiple directions to evenly coat the agar plate in the top agar mixture.
- The agar plate is then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plate was flipped over and placed in the proper incubator to be incubated for 24-48 hours at a temperature of 29˚C .
Results:
– Following a 24 hour period, the sample was taken out of the incubator and signs of plaque formations were located on the upper left and center of the plate. The larger “plaque” in the upper center can be ignored as a bubble was accidentally formed during the final steps of the plaque assay during step 7. Three photos will be attached below:
Plaque Assay 24 hour for sample 6 pt 1 Plaque Assay 24 hour for sample 6 pt 2 Plaque Assay 24 hour for sample 6 pt 3
– Following a 48 hour period, the sample was taken out of the incubator and the agar plate showed small formations of plaques as shown in the picture below, deeming the experiment successful.
Plaque Assay 48 hour for sample 6
Conclusions and Next Steps:
– After collecting the plate, it was wraped tightly in parafilm to store in the refrigerator at 4˚C to prevent contamination. This plate will be used to begin the serial dilutions of the phage.
Amplification
Title: First Round of Serial Dilution to Purify Phage
Date: 09/25/2023
Redo: No
Sample: #6
Purpose: To prepare liquid phage samples of decreasing concentrations.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin the first round of Serial Dilutions to purify the phage
- Supplies:
- Phage buffer
- Samples requiring diluting (e.g. plaques from sample #6)
- Microcentrifuge tube
- Bunsen Burner
- To set up the 10-fold serial dilutions, eight microcentrifuge tubes were racked and each was labeled in order from 10-1, 10-2, 10-3,….10-6. (Note: Textbook notes that a total of 8 microcentrifuge tubes should be labeled and racked in order of 10-1, 10-2, 10-3,….10-8. but since only 10-1, 10-2, 10-3,….10-6 is required, we will only rack a total of six microcentrifuge tubes.)
- Using the correct micropippet, 90μl of phage buffer is added to each microcentrifuge tube. (Note: the bunsen burner was lit and the supplies were put near the burner to make sure the supplies will stay sterile under the sterilization cone provided by the flame)
- To perform the 10-fold serial dilutions, the micropippetor was used to collect the phages from the one of the plaques formed on the plate without touching any of the bacteria covered surfaces on the agar plate. (Note: only the tip of the micropipettor needs to be covered in the phages from the agar plate instead of 10μl of phages)
- Once the phages were collected from the agar plate, the micropipettor was used to dispense the phages into the “10-1” microcentrifuge tube and was vortexed well to properlly mix the phages with the buffer.
- The solution in this “10-1” microcentrifuge tube contains 1/10th the number of phage particles as the undiluted phage sample. It is also referred to as a 1:10 dilution.
- Make sure to use a clean pipette tip for each transfer and pipette carefully, vortexing the sample well before making each dilution. Otherwise, there will not be accurate 10-fold dilutions.
5. 10 μl of the “10 -1” sample is then transferred to the “10-2” tube and is vortexed well.
- This solution now containes 1/100th as many phage particles as the undiluted sample.
6. Each successive dilution is continued until the last tube (10-6 ) is reached.
Results:
Once each microcentrifuge tube was filled with the successive dilutions, each tube contained a clear fluid that was filled to approximately 100μl of the sample-buffer mix.
Conclusions and Next Steps:
After the serial dilution was performed, the table was prepared for aseptic work and supplies were gathered to begin the plaque assay of each dilution.
————————————————————————————————————-
Title: Plaque Assay for Purification of First Round of Serial Dilutions
Date: 09/25/2023
Redo: No
Sample: #6
Purpose: To generate well-isolated plaques for second round of serial dilutions.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin the first round of Serial Dilutions to purify the phage
- Supplies:
- Phage Samples for purification
- Microcentrifuge tubes
- Host Bacteria
- Agar plates
- Top agar
- Micropipettes
- Since this procedure is the same as a regular plaque assay, this procedure will follow the same steps as plaque assay.
- Once the tubes were capped following serial dilutions, six test tubes containing 10 μl host bacterial cultures was collected as well as an agar plate.
- Using a micropipettor, the 10μl of the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria M. foliorum and was gently mixed by tapping the tube.
- This process is repeated until each of the six bacterial culture tubes were filled with their respective diluted sample.
- Once the tubes were mixed, the tubes were left undisturbed for around 5 minutes to allow for attachment.
- 6 agar plates are then labeled accordingly with each dilution and set on the table for future use.
- A bottle of molten top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacterial culture tubes.
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate, making sure to avoid making or sucking up bubbles, as they can be confused for plaques on the agar plates. We then immediately discarded the pipette.
- The agar plate is gently and quickly tilted in multiple directions to give the agar plate an even and level coat of the molten top agar.
- Steps 8-10 are then repeated six times until each respective plate is coated in molten top agar, with each plate containing the reference to the dilution of the sample used. (ex: one plate will contain the “10 -1” diluted sample mixed top agar and so on.)
- The agar plates are then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plates were flipped over and placed in the proper incubator to be incubated for 48 hours at a temperature of (29°C).
Results:
After 24 hours of incubation, plates “10 -1” – “10 -3” showed signs of plaques, while “10 -4”- “10 -6” did not show signs of plaques, the plates were placed back into the incubator and left for another 24 hours for further plaque development. The pictures are provided below:
24 Hour Serial Dilution Plaque Assay for sample 6
After 48 hours of incubation, plates “10 -1” – “10 -4” showed signs of plaque development, while plates “10 -5” – “10 -6” showed no sings of plaque development as shown in the picture provided below.
48 Hour Serial Dilution Plaque Assay of sample 6
Conclusions and Next Steps:
To further isolate the bacteriophage, a second round of serial dilutions will be performed using the plaques found on the plate containing “10 -4” dilution as the plate has the least concentrated number of phages. The remaining five plates were discarded as they will no longer be used.
Title: Second Round of Serial Dilution to Purify Phage
Date: 09/27/2023
Redo: No
Sample: #6
Purpose: To prepare liquid phage samples of decreasing concentrations..
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin the first round of Serial Dilutions to purify the phage
- Supplies:
- Phage buffer
- Samples requiring diluting (e.g. plaques from plate containing “10 -4” dilution from previous serial dilution plaque assay)
- Microcentrifuge tube
- Bunsen Burner
- To set up the 10-fold serial dilutions, eight microcentrifuge tubes were racked and each was labeled in order from 10-1, 10-2, 10-3,….10-6. (Note: Textbook notes that a total of 8 microcentrifuge tubes should be labeled and racked in order of 10-1, 10-2, 10-3,….10-8. but since only 10-1, 10-2, 10-3,….10-6 is required, we will only rack a total of six microcentrifuge tubes.)
- Using the correct micropippet, 90μl of phage buffer is added to each microcentrifuge tube. (Note: the bunsen burner was lit and the supplies were put near the burner to make sure the supplies will stay sterile under the sterilization cone provided by the flame)
- To perform the 10-fold serial dilutions, the micropippetor was used to collect the phages from the one of the plaques formed on the plate without touching any of the bacteria covered surfaces on the agar plate. (Note: only the tip of the micropipettor needs to be covered in the phages from the agar plate instead of 10μl of phages)
- Once the phages were collected from the agar plate, the micropipettor was used to dispense the phages into the “10-1” microcentrifuge tube and was vortexed well to properlly mix the phages with the buffer.
- The solution in this “10-1” microcentrifuge tube contains 1/10th the number of phage particles as the undiluted phage sample. It is also referred to as a 1:10 dilution.
- Make sure to use a clean pipette tip for each transfer and pipette carefully, vortexing the sample well before making each dilution. Otherwise, there will not be accurate 10-fold dilutions.
5. 10 μl of the “10 -1” sample is then transferred to the “10-2” tube and is vortexed well.
- This solution now containes 1/100th as many phage particles as the undiluted sample.
6. Each successive dilution is continued until the last tube (10-6 ) is reached.
Results:
Once each microcentrifuge tube was filled with the successive dilutions, each tube contained a clear fluid that was filled to approximately 100μl of the sample-buffer mix..
Conclusions and Next Steps:
After the serial dilution was performed, the table was prepared for aseptic work and supplies were gathered to begin the plaque assay of each dilution.
Title: Plaque Assay for Purification of Second Round of Serial Dilutions
Date: 09/27/2023
Redo: No
Sample: #6
Purpose: To generate well-isolated plaques for second round of serial dilutions.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin the first round of Serial Dilutions to purify the phage
- Supplies:
- Phage Samples for purification
- Microcentrifuge tubes
- Host Bacteria
- Agar plates
- Top agar
- Micropipettes
- Since this procedure is the same as a regular plaque assay, this procedure will follow the same steps as plaque assay.
- Once the tubes were capped following serial dilutions, six test tubes containing 10 μl host bacterial cultures was collected as well as an agar plate.
- Using a micropipettor, the 10μl of the sample in the microcentrifuge tube was transferred into the tube containing the host bacteria M. foliorum and was gently mixed by tapping the tube.
- This process is repeated until each of the six bacterial culture tubes were filled with their respective diluted sample.
- Once the tubes were mixed, the tubes were left undisturbed for around 5 minutes to allow for attachment.
- 6 agar plates are then labeled accordingly with each dilution and set on the table for future use.
- A bottle of molten top agar was removed from a 55°C bath (Note: the top agar needs to stay at 55°C for as long as possible before use as to prevent it from solidifying prematurely).
- A sterile 5ml pipette is used to aseptically transfer 3 ml of top agar to the bacterial culture tubes.
- The mixture is immediately aspirated back into the pipette and transferred into the appropriate plate, making sure to avoid making or sucking up bubbles, as they can be confused for plaques on the agar plates. We then immediately discarded the pipette.
- The agar plate is gently and quickly tilted in multiple directions to give the agar plate an even and level coat of the molten top agar.
- Steps 8-10 are then repeated six times until each respective plate is coated in molten top agar, with each plate containing the reference to the dilution of the sample used. (ex: one plate will contain the “10 -1” diluted sample mixed top agar and so on.)
- The agar plates are then left to sit undisturbed for 20 minutes.
- Once the 20 minutes had passed, the plates were flipped over and placed in the proper incubator to be incubated for 48 hours at a temperature of (29°C).
Results:
After 24 hours of incubation, plates “10 -1” – “10 -4” showed signs of plaque formation, with “10 -1” showing the most and gradually decreasing in number until reaching “10 -4”, which showed only 2 plaques (Note: a bundle of what seems to be plaques are shown near the center of plate “10 -3”, but is actually a bundle of bubbles). Plates “10 -5”-“10 -6” showed no signs of plaque formation, with plate “10 -6” showing what seems like a plaque, but is actually a bubble formation. a picture is taken of the plates, which is shown below, and the plates are placed back in the incubator for another 24 hours.
Round 2 Serial Dilution Plaque Assay 24 Hour Period
After 48 hours of incubation, plates “10 -1” – “10 -5” showed signs of plaque formation, with “10 -1” showing more plaques than before, and gradually decreasing until reaching plate “10 -5”, which shows only 1 plaque formation, which is shown behind the “f” of the labeled plate. As mentioned above, plate “10 -6” showed no signs of plaque formation, with what seems like plaques only being bubble formations from the plaque assay procedure. Pictures are taken of each plate and parafilm is placed on each plate to be put in the fridge for further experimentation.
48 Hour Second Round of Serial Dilution Plaque Assay
Conclusions and Next Steps:
After the second round of serial dilutions, it was shown that plate “10 -5” showed the least amount of plaque formations, which therefore has the lowest concentration of bacteriophages and will be used for further experimentation for the procedure.
Title: Collecting Low Volume Plate Lysates
Date: 10/2/2023
Redo: No
Sample: #6
Purpose: To generate a highly concentrated liquid phage sample.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin collection of plate lysates
- Supplies:
- Webbed plate(s) with clonal phage population
- Phage Buffer (8 mL/plate)
- 0.22 μm filter
- 5 ml syringe
- 15 ml sterile conical tube for lysate storage
- Bunsen burner
- Prior to start of procedure, the agar plate containing (10-1) from the previous plaque assay of second round of serial dilutions was collected since it was identified as a webbed plate, which is required to do the plate lysate procedure.
- Once the plate was collected, 8mL of sterile phage buffer was applied to the webbed plate, swirlled to cover the plate in phage buffer without splashing, and was left undisturbed for around 6 hours at room temperature.
- After the 6 hour period had passed, the lid of the plate was removed and placed on the bench. The plate was tilted by placing one edge of the plate on the lid, allowing the lysate to pool to one side. (Note: a bunsen burner was lit and the materials were placed near the burner to be covered by the sterile field created by the burner)
- A 0.22 μm filter was perepared by opening the packaging, but the filter was not removed yet.
- Using a 5mL syringe, the lysate was aspirated (sucked up) from the plate.
- once the lysate was collected, the filter was carefully attached to the syringe and the syringe plunger was slowly depressed to dispense the filtrate into a 15 mL conical tube.
- Since some lysate was still remaining in the plate, the filter was carefully removed as to not contaminate the filter, and the remaining lysate was aspirated and filtered into the conical tube.
- After dispensing the filtrate into the tube, the tube filled to the 7mL mark, and was labeled accordingly and was stored into a fridge at 4 °C for further procedures requiring lysate.
Results:
Once the tube was labelled accordingly and the amount of filtrate was measured (7mL), the tube was stored in the fridge for the next steps of procedure.
Conclusions and Next Steps:
Following the experiment, the next steps to be taken are to calculate the titer of the lysate filtrate using Full Plate Titer protocol.
Title: Full Plate Titer
Date: 10/4/2023
Redo: No
Sample: #6
Purpose: To determine the concentration of phage particles in a lysate by using a plaque assay.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin full plate titer
- Supplies:
-
- Liquid phage sample requiring titering
- Phage buffer
- Agar plates for plating dilutions
- Top agar, molten (between 55 – 60 ˚C)
- Host bacteria
- Microcentrifuge tubes
- 5 ml serological pipettes
- Bunsen Burner
- The 15mL labelled conical tube containg the lysate filtrate was removed from the refrigerator and was set aside to perform serial dilutions of the filtrate.
- Using the lysate filtrate, a plaque assay procedure is followed using the same steps as used in Plaque Assay for Purification of Second Round of Serial Dilutions but with only four agar plates to dilute the sample to concentrations “10 -1” – “10 -4”.
- The next day, the plates were checked to confirm that the dilutions were valid (i.e., a 10-fold decrease in plaques).
- Once the plate containing the concentration “10 -4” was collected, the titer was calculated using the formula: Titer (pfu/ml) = (# pfu/ volume used in μl) x (103 μl/ml) x dilution factor*
Results:
Once the plates were collected after 24-48 hour incubation following the plaque assay of the lysate filtrate, only plate “10 -4” seemed to have any plaques formed on the plate, as the “10 -1” – “10 -3” seemed to have been possibly contaminated. The pictures of the plates are shown below:
Plaque assay 24 hour for lysate dilutions
Plaque assay 48 hour for lysate dilutions
Since there were to many plaques to correctly count on plate “10 -4”, the titer would be impossible to calculate, deeming the experiment unsuccessful.
Conclusions and Next Steps:
Following the unsuccessful experiment, the next steps will be to make a second attempt of full plate titer using the lysate filtrate to get a proper amount of plaques to calculate the titer of the sample.
Title: Full Plate Titer
Date: 10/6/2023
Redo: Yes
Sample: #6
Purpose: To determine the concentration of phage particles in a lysate by using a plaque assay.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin full plate titer
- Supplies:
-
- Liquid phage sample requiring titering
- Phage buffer
- Agar plates for plating dilutions
- Top agar, molten (between 55 – 60 ˚C)
- Host bacteria
- Microcentrifuge tubes
- 5 ml serological pipettes
- Bunsen Burner
- The 15mL labelled conical tube containg the lysate filtrate was removed from the refrigerator and was set aside to perform serial dilutions of the filtrate.
- Using the lysate filtrate, serial dilutions a plaque assay procedure is followed using the same steps as used in Plaque Assay for Purification of Second Round of Serial Dilutions but with only four agar plates to dilute the sample to concentrations “10 -1” – “10 -4”.
- The next day, the plates were checked to confirm that the dilutions were valid (i.e., a 10-fold decrease in plaques).
- Once the plate containing the concentration “10 -4” was collected, the titer was calculated using the formula: Titer (pfu/ml) = (# pfu/ volume used in μl) x (103 μl/ml) x dilution factor*
Results:
Once the plates were collected after 24-48 hour incubation following the plaque assay of the lysate filtrate, only plate “10 -4” seemed to have any plaques formed on the plate, as the “10 -1” – “10 -3” seemed to have been possibly contaminated. The pictures of the plates are shown below:
Plaque assay 24 hour for lysate dilutions
Plaque assay 48 hour for lysate dilutions
Since there were to many plaques to correctly count on plate “10 -4”, the titer would be impossible to calculate, deeming the experiment unsuccessful.
Conclusions and Next Steps:
Following the unsuccessful experiment, the next steps will be to make a second attempt of full plate titer using the lysate filtrate to get a proper amount of plaques to calculate the titer of the sample. However, we will be using 3 10-4 serial dilutions for our next 3 plates as the 10-4 plate showed the most prominent bacteria growth.
Title: Full Plate Titer
Date: 10/6/2023
Redo: Yes
Sample: #6
Purpose: To determine the concentration of phage particles in a lysate by using a plaque assay.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin full plate titer
- Supplies:
-
- Liquid phage sample requiring titering
- Phage buffer
- Agar plates for plating dilutions
- Top agar, molten (between 55 – 60 ˚C)
- Host bacteria
- Microcentrifuge tubes
- 5 ml serological pipettes
- Bunsen Burner
- The 15mL labeled conical tube containing the lysate filtrate was removed from the refrigerator and was set aside to perform serial dilutions of the filtrate.
- Using the lysate filtrate, we began by setting up the procedure for Serial Dilutions. We proceeded, following the steps for this procedure, but instead made three separate 10-4 microcentrifuge tubes to use for our agar plates, as those would be the three we would be using to cultivate phage growth.
- Using the serial dilutions, a plaque assay procedure is followed using the same steps as used in Plaque Assay for Purification of the Second Round of Serial Dilutions but with only three agar plates to dilute the sample to concentrations all diluted 10 -4
- The next day, the plates were checked to confirm that the dilutions were valid (i.e., a 10-fold decrease in plaques).
Results:
Once the plates were collected after 24-48 hour incubation following the plaque assay of the lysate filtrate, all plates showed signs of contamination, thus proving unusable in the steps following.
Plaque-assay-24-hour-for-redo-lysate-dilutions
Conclusions and Next Steps:
Following the unsuccessful experiment, these steps will be repeated to get a full plate titer without any contamination that can be used to calculate the titer.
Title: Full Plate Titer
Date: 10/11/2023
Redo: Yes
Sample: #6
Purpose: To determine the concentration of phage particles in a lysate by using a plaque assay.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin full plate titer
- Supplies:
-
- Liquid phage sample requiring titering
- Phage buffer
- Agar plates for plating dilutions
- Top agar, molten (between 55 – 60 ˚C)
- Host bacteria
- Microcentrifuge tubes
- 5 ml serological pipettes
- Bunsen Burner
- The 15mL labeled conical tube containing the lysate filtrate was removed from the refrigerator and was set aside to perform serial dilutions of the filtrate.
- Using the lysate filtrate, we began by setting up the procedure for Serial Dilutions. We proceeded, following the steps for this procedure, but instead made three separate 10-4 microcentrifuge tubes to use for our agar plates, as those would be the three we would be using to cultivate phage growth.
- Using the serial dilutions, a plaque assay procedure is followed using the same steps as used in Plaque Assay for Purification of the Second Round of Serial Dilutions but with only three agar plates to dilute the sample to concentrations all diluted 10 -4
- The next day, the plates were checked to confirm that the dilutions were valid (i.e., a 10-fold decrease in plaques).
Results:
Once the plates were collected after 24-48 hour incubation following the plaque assay of the lysate filtrate, all plates showed signs of contamination, thus proving unusable in the steps following.
48-hour-plaque-assay-redo-lysate-dilutions
Note:
Conclusions and Next Steps:
Following discussion with Dr. Edwards, due to multiple repeated attempts of full plate titers showing possible signs of contamination, Dr. Edwards checked the results of this full plate titer attempt and said that its possible that it may not be contaminated and to continue with the next steps. The nexts steps to take is to begin the high volume lysate procedure.
Title: Collecting High Volume Plate Lysates
Date: 10/16/23
Redo: No
Sample: #6
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin collection of plate lysates
- Supplies:
- Webbed plate(s) with clonal phage population
- Phage Buffer (8 mL/plate)
- 0.22 μm filter
- 5 ml syringe
- 15 ml sterile conical tube for lysate storage
- Bunsen burner
- Prior to start of procedure, the four agar plates containing (10-4) from the previous plaque assay of low volume plate lysate titers was collected since they were identified as a webbed plate, which is required to do the plate lysate procedure.
- Once the plates were collected, 8mL of sterile phage buffer was applied each of webbed plates, swirlled to cover the plate in phage buffer without splashing, and was left undisturbed for around 3 hours at room temperature.
- After the 3 hour period had passed, the lid of the plate was removed and placed on the bench. The plates were tilted by placing one edge of the plate on the lid, allowing the lysate to pool to one side. (Note: a bunsen burner was lit and the materials were placed near the burner to be covered by the sterile field created by the burner)
- A 0.22 μm filter was perepared by opening the packaging, but the filter was not removed yet.
- Using a 5mL syringe, the lysate was aspirated (sucked up) from the plate.
- once the lysate was collected, the filter was carefully attached to the syringe and the syringe plunger was slowly depressed to dispense the filtrate into a 50 mL conical tube.
- Since some lysate was still remaining in the plate, the filter was carefully removed as to not contaminate the filter, and the remaining lysate was aspirated and filtered into the conical tube.
- Approximately three 0.22 μm filters were used and one vacuum filtration kit was used to filter all four agar plates containing the lysate.
- After dispensing the filtrate into the tube, the tube filled to the 20mL mark, and was labeled accordingly and was stored into a fridge at 4 °C for further procedures requiring lysate.
Results:
Once the tube was labelled accordingly and the amount of filtrate was measured (20mL), the tube was stored in the fridge for the next steps of procedure.
Conclusions and Next Steps:
Following the experiment, the next steps to be taken are to calculate the titer of the high lysate filtrate using Full Plate Titer protocol.
Title: Full Plate Titer
Date: 10/18/2023
Redo: No
Sample: #6
Purpose: To determine the concentration of phage particles in a lysate by using a plaque assay.
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled to begin full plate titer
- Supplies:
-
- Liquid phage sample requiring titering
- Phage buffer
- Agar plates for plating dilutions
- Top agar, molten (between 55 – 60 ˚C)
- Host bacteria
- Microcentrifuge tubes
- 5 ml serological pipettes
- Bunsen Burner
- The 50mL labelled conical tube containg the lysate filtrate was removed from the refrigerator and was set aside to perform serial dilutions of the filtrate.
- Using the lysate filtrate, a plaque assay procedure is followed using the same steps as used in Plaque Assay for Purification of Second Round of Serial Dilutions but with only four agar plates to dilute the sample to concentrations “10 -1” – “10 -4”.
NOTE: Since plates “10 -1” – “10 -4” had too many plauqes to properly count, an extra 3 plates ranging from “10 -5” – “10 -7” were made using plaque assay and serial dilutions to collect a plate that would contain a countable number of plaques.
- The next day, the plates were checked to confirm that the dilutions were valid (i.e., a 10-fold decrease in plaques).
- Once the plates were collected, the agar plate with a countable amount of plaques (“10 -7 “dilution) was used to calculate the titer of the sample using the formula: Titer (pfu/ml) = (# pfu/ volume used in μl) x (103 μl/ml) x dilution factor*
Results:
The pictures below showed the result of the Plaque Assay’s and Serial dilutions:
Plaque Assay 48 Hour for High Titer Dilutions , Plaque Assay 48 Hour for Extra High Titer Dilutions
Following the plaque assay and serial dilutions of plates “10 -1” – “10 -7”, the plate that contained a countable number was used to calculate the full plate titer, that being plate “10 -7” with a total of 15 plaques. With the number of plaques counted, the titer was calculated to be 1.5 x 1010 pfu/ml using the formula Titer (pfu/ml) = (# pfu/ volume used in μl) x (103 μl/ml) x dilution factor*
Conclusion and Next Steps:
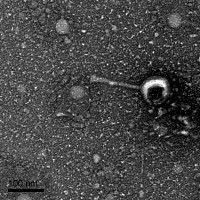
Once the titer was calculated, the plates were parafilmed and placed in the fridge for possible future use and the next steps were to conduct a TEM and stain of the sample.
DNA Extraction
Title: DNA Extraction
Date: 11/06/23
Redo: No
Sample: #6
Purpose: To isolate genomic DNA from phage.
Notes:
- Prior to start of procedure, the bench was cleaned and supplies were assembled to begin DNA Extraction
- Supplies:
- 1 ml phage lysate (titer ≥ 5 x 109 pfu/ml)
- Nuclease mix
- EDTA (0.5M) (Optional)
- Proteinase K (20 mg/mL) and SDS (10%) (Optional)
- 2mL DNA clean-up resin (Prometa Wizard DNA Clean-Up Kit)
- 2 DNA clean-up columns (Promega Wizard DNA Clean-Up Kit)
- 3 ml syringes
- 6 ml 80 % isopropanol, freshly prepared
- ddH2O pre-warmed (90 °C)
- Degrade bacterial DNA/RNA in high-titer phage lysate.
- 1 mL of the high volume phage lysate was aseptically transferred into a microcentrifuge tube.
- While wearing gloves and working in the fume hood designated area, 5µl of nuclease mix was carefully added to the lysate. Important: The enzymes (RNase in particular) are very stable and can persist and contaminate equipment and supplies throughout the laboratory. Take precautions to keep and use them in the designated area.
- The nuclease was mixed gently and thouroughly through repeated inversions. Note: do not vortex the mixture!
- 15 µl EDTA was added to the nuclease-treated lysate and was mixed gently.
- The EDTA will inactivate the nucleases by chelating, or binding, divalent cations required by the nucleases for activity.
- After mixing, 0.5 µl Proteinase K and 50 µl SDS was added to the mixture and was mixed gently.
- The Proteinase K is added to degrade the nucleases added in Step 2. SDS stimulates the activity of Proteinase K.
- The mixture was then incubated at 37 °C for 10 minutes. while the mixture is incubating, the gloves are removed and discarded prior to returning to the bench for the next steps.
- Denature the protein capsid to release phage DNA.
- A fresh pair of gloves was put on prior to next steps.
- 2 ml of DNA clean-up resin was aded to a 15mL conical tube. Important: The resin contains guanidinium thiocyanate, a chemical that denatures proteins. Do not get it on your skin!
- The nuclease-treated phage lysate was then transferred from the microcentrifuge tube to the 15 ml conical tube containing resin.
- The solution was mixed by gently inverting the tube repeatedly for 2 minutes.
- Isolate the phage genomic DNA.
- Two Wizard Kit colums are collected, as well as two syringes.
- The plungers from the two syringes are removed and the column was attached to each syringe barrel.
- The next steps were followed for each column at the same time.
- The column and syringe barrel were set on a new microcentrifuge tube.
- 1.5 ml of phage DNA/resin solution was transferred to the column using a pipette. The empty 15 mL conical tube was not discarded at this time.
- a plunger was inserted into a syring and all the liquid was carefully pushed through, collecting the flow-through in the used 15 mL conical tube. Important: The DNA is bound to the polymer beads that pack into the column as the liquid is pushed through. It is VERY important to maintain a firm, gentle, unrelenting, and even pressure on the syringe. Do not let the plunger pop out of the syringe barrel because releasing the vacuum will ruin the column.
- Once the liquid was expelled, pressure was maintained on the plunger as the residual liquid was dried by touching the tip of the column on a paper towel.
- The column was unscrewed from the syringe barrel before releasing the plunger the column was set into a clean microcentrifuge tube.
- The plunger was then removed from the syringe barrel, and then, the column was reattached to the syringe barrel.
- Wash the salts from the DNA (now in the column) with the following steps for each column:
- 2 ml 80 % isopropanol was added to each syringe barrel/column and the liquid was pushed through the column, repeating steps 3(3)-3(5).
- This process is repeated twice for a total of three isopropanol washes.
- Remove residual isopropanol.
- Each column was placed in a fresh 1.5mL microcentrifuge tube and was spun at 10,000 × g for 5 minutes.
- The column will prevent the microfuge tube lids from closing. Arrange the open tubes in the centrifuge so that the lids point toward the center of the rotor.
- following the centrifuge, the columns are placed into new 1.5mL microcentrifuge tubes and are spun at 10,000 x g for 1 additional minute to remove any residual isopropanol.
- The last traces of isopropanol were evaporated by removing the columns from the microcentrifuge tubes and placing the columns directly in a 90°C heating block for 60 seconds.
Important: Leaving the columns in the heat block for more than 1 minute can lead to DNA damage.
- Each column was placed in a fresh 1.5mL microcentrifuge tube and was spun at 10,000 × g for 5 minutes.
- The phage DNA was eluted from the column.
- Each column was placed in a clean microcentrifuge tube and 50 μl of 90 °C sterile ddH2O was directly applied to each column. Important: Keep the ddH2O in the heating block so that it remains at 90 °C.
- The columns are incubated for 1 minute at room temperature.
- The columns are then spun at 10,000 × g for 1 minute in a microcentrifuge.
- The products from both microcentrifuge tubes are combined into one tube; this being the eluted phae DNA.
- Determine the concentration of your phage DNA.
- A spectrophotometer (fluorimeter, or Nanodrop) and a protocol from your instructor was used to quantify the phage DNA.
- The phage was then placed at 4 °C for short-term storage (1–2 weeks) or at -20 °C for long-term storage.
Results:
Following the experiment, the microcentrifuge tube contained a clear solution, once placed under a spectrophotometer, the following picture was observed:
Conclusions and Next Steps:
Due to properties of the scan, the extraction showed unsatisfactory results. The next steps are to repeat the DNA Extraction procedure using another method.
Title: DNA Extraction
Date: ##/###/##
Redo: Yes/No
Sample: #
Purpose: ###.
Notes:
Results:
###.
Conclusions and Next Steps:
###.
Characterization
Title: Setting Up Restriction Enzyme Digests
Date: 11/14/2023
Redo: No
Sample: #6
Purpose: To cut the phage genome into multiple fragments based on its DNA sequence.
Notes:
- Prior to start of procedure, the bench was cleaned and supplies were assembled to begin Setting up Restriction Enzyme Digests.
-
Supplies:
- Phage DNA
- 37 °C water bath
- 65 °C heat block
- Microcentrifuge tubes
- Restriction enzymes λ-HindIII, uncut, HaeIII, Msel, Nspl, SacII and SaII with buffers
- Procedure:
- Prepare genomic DNA.
- The DNA was taken out from the freezer and left to thaw out for around 10 minutes.
- The DNA was gently mixed by flicking the closed microcentrifuge tube with a finger.
- Once mixed, the tube was incubated at 65 °C for 10 minutes, and then was quick spined in a microcentrifuge for 30 seconds to move the liquid to the bottom of the tube, and was then quickly placed on ice.
- The volume of the DNA sample was calculated to obtain 0.5 µg of DNA using the concentration of the sample (1374ng/µL), which showed that a total of 6µL of DNA would be used to set up the restriction digest reactions.
-
- Set up restriction enzyme digest reactions.
- Set up a reaction in a microcentrifuge tube for each enzyme according to the table below for a total of 7 microcentrifuge tubes:
| Solution | Volume |
| Sterile ddH2O | 20.5µL |
| Reaction Buffer | 2.5µL |
| Restriction Enzyme | 1µL |
| Phage Genomic DNA | 1µL |
Important: The phage DNA was added last to prevent contamination of the enzyme stocks.
- In the first microcentrifuge tube labeled “λ-HindIII”, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of the restriction enzyme was added to the tube, and lastly 1µL of the λ-HindIII was added, as it contains its own DNA and will be used as a ladder.
- In the second microcentrifuge tube labeled “uncut”, 24µL of Sterile ddH2O was added, and then 1µL of phage genomic DNA was added.
- In the third microcentrifuge tube labeled “HaeIII”, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of HaeIII was added to the tube, and lastly 1µL of phage genomic DNA was added.
- In the fourth microcentrifuge tube labeled “Msel”, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of Msel was added to the tube,and lastly 1µL of phage genomic DNA was added.
- In the fifth microcentrifuge tube labeled “Nspl”, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of Nspl was added to the tube, and lastly 1µL of phage genomic DNA was added.
- In the sixth microcentrifuge tube labeled “SacII “, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of SacII was added to the tube, and lastly 1µL of phage genomic DNA was added.
- In the seventh microcentrifuge tube labeled “SaII “, 20.5µL of Sterile ddH2O was added, then 2.5µL of the reaction buffer was added, next 1µL of SaII was added to the tube, and lastly 1µL of phage genomic DNA was added.
- During steps 1-7, after each tube was completed, the microcentrifuge tube was centrifuged for around 15 seconds to move all of the liquid to the bottom of the tube.
- Once all seven of the tubes were completed, they were all incubated at 37 °C for 15 minutes.
- After the tubes had incubated, they were taken out and each tube was spun in a microcentrifuge for 30 seconds to move the liquid to the bottom of the tube and were stored at -20 °C until ready to use.
Results:
Once each tube was finished centrifuging, each tube contained a clear fluid. These tubes were then placed in a freezer to store until later use.
Conclusions and Next Steps:
Once the tubes were placed in the freezer, the next steps to take were to cast the agarose gels.
Title: Casting Agarose Gels
Date: 11/14/2023
Redo: No
Sample: #6
Purpose: To prepare an agarose gel for electrophoresis
Notes:
- Prior to start of procedure, the bench was cleaned and supplies were assembled to begin Setting up Casting Agarose Gels.
-
Supplies:
- Agarose
- 1X TBE running buffer (TBE = 0.089 M Tris-Boric Acid, 0.002 M-EDTA)
- DNA dye (ethidium bromide or alternative)
- Erlenmeyer flask
- Electrophoresis apparatus and power supply
- Procedure:
- Pour a 0.8 % (w/v) agarose gel.
- A gel apparatus was set up to prepare for agarose gel placement.
- approximately 0.33 g of agarose powder was transferred into an Erlenmeyer flask
- 40mL of 1X TBE running buffer was then added to the Erlenmeyer flask and was swirled to mix.
- Once mixed for a few seconds, a paper towel was then plugged into the top of the flask to prevent any spillage and the flask was placed into a microwave for 1 minute to boil.
- Once the minute had passed, using a heat-resistant glove, the flask was carefully removed from the microwave and left to cool at room temperature.
- After the flask had cooled for an appropriate amount of time, gloves were then put on and the gel was added into the Electrophoresis apparatus appropriately.
- The Gel was then left to cool for 30 minutes with the comb left inside to allow for the formation of wells within the gel.
- Once the gel had solidified, the comb was carefully removed by slowly pulling it straight up, once the comb was removed, the gel platform was gently lifted out of the casting tray.
- The platform with the solidified gel was placed into the gel box with the wells at the (-) cathode end of the box, where the black lead is connected.
- Then 1X TBE buffer was poured into the gel apparatus until the gel is submerged by ~ 1/4 inch of buffer.
- Load your samples and run the gel according to the Gel Electrophoresis protocol
Results:
Once the buffer was poured, the apparatus contained the gel with the wells at the (-) cathode end of the box and the buffer slightly above the gel.
Conclusions and Next Steps:
Once the apparatus was set up, the Gel Electrophoresis protocol can be started.
Title: Gel Electrophoresis of Restriction Enzyme Digests
Date: 11/14/2023
Redo: No
Sample: #6
Purpose: To separate DNA fragments via agarose gel electrophoresis
Notes:
- Prior to start of procedure, the bench was cleaned and supplies were assembled to begin Setting up Gel Electrophoresis of Restriction Enzyme Digests.
-
Supplies:
- Pre-poured agarose gel
- DNA loading dye
- Electrophoresis apparatus and power supply
- DNA cut with restriction enzymes
- DNA ladder
- Small tupperware box
- DI water
- Procedure:
- Set up the gel electrophoresis equipment with a gel prepared according to the Casting Agarose Gels protocol.
- Wearing gloves, the gel was oriented in a way that the wells are closes to the cathode (black electrode).
-
- Prepare your restriction enzyme digest samples for electrophoresis.
- 5 of μl of concentrated purple loading dye was added to each of the 25 µl restriction enzyme samples prepared in the “Setting up Restriction Enzyme Digests” protocol.
- The samples were placed at 65 °C in a heat block for 5 minutes and were then immediately placed on ice to cool. And were then spun in a microcentrifuge for ~15 seconds.
-
- Load the gel in the following order:
λ-HindIII Uncut DNA HaeIII MseI NspI SacII SaII
- Load the gel in the following order:
- The gel was carefully loaded with the proper colume of DNA ladder.
- Using a fresh tip of the micropipettor for each sample, 20 μl was pipetted of each RE reaction into the wells in the order above.
- Holding the pipette in both hands, both elbows were placed on either side of the gel apparatus.
- The eyes were situated directly above the wells to make the wells easier to see.
- The pipette tip was placed directly above the well, just below the surface of the buffer. Do not try to get the pipette tip into the well, or you might puncture the bottom of the gel.
- The pipette plunger was slowly depressed, allowing the solution to slowly sink into the well.
- The pipette was removed from the gel before releasing the plunger.
- The electrodes were then plugged into the appropriate locations on the power supply. The power supply was turned on and the voltage was set to 100 V.
- Important: Remember, DNA runs toward the RED electrode!
- For approximately 1 and a half hours, the gel ran until the purple dye had migrated at least 3.5 inches from the well.
- Once the dye reached the destination, the power supply was tuned off and the gel was gently taken out of the apparatus and was placed into a tupperware box and was filled with buffer until the gel was covered by 1/4 inch of buffer solution and 4μl of EtBr for staining.
- The box was then placed onto a gel rocker for 15 minutes.
- After the 15 minutes had passed, the rocker was turned off and the box was emptied of the buffer, without emptying the gel by gently holding the gel in place. Then the tupperware box was filled with DI water until the gel was covered by 1/4 inch of DI water and was placed onto the rocker for another 15 minutes.
- After the 15 minute period, the box was taken off the rocker and was again emptied without removing the gel, and the box was again filled with DI water until he gel was covered by 1/4 inch of DI water, which was placed onto the rocker for an additional 15 minutes.
- Following the 15 minute period, the box was taken off the rocker and the DI water was emptied from the box, leaving the gel inside the box to photograph.
Results:
Following the procedure, the gel was placed onto a UV light box to view the DNA fragments as shown by the photo below:
Conclusions and Next Steps:
The photo shows that the phage sample contains approximately ~22,000 bp when compared to the ladder (λ-HindIII).
Title: Mounting Phage Samples for TEM and Staining with Uranyl Acetate
Date: 10/25/2023
Redo: No
Sample: #6
Purpose: To prepare your phage sample for viewing with a transmission electron microscope
Notes:
- Prior to procedure, the bench was cleaned and supplies were assembled
1. Approximately 1 mL of the high-titer lysate is transferred into a sterile microcentrifuge tube, which is placed in a centrifuge at 4°C and is set at top speed for 1 hour in order to pellet the phage particles to the bottom of the tube. (the microcetrifuge tube is balanced using a microcentrifuge tube filled with water across from it)
2. Once the hour had passed, using a micropipettor, as much of the supernatant as possible was removed without disrupting the pellet, leaving as little liquid as possible without dislodging the pellet. (the pellet was identified as a tiny yellow pellet at the bottom of the tube)
3. The pellet is resuspended in a 100μl of phage buffer and was placed back in the centrifuge at 4°C for another 30 minutes to mix.
4. After the 30 minute period, the tube was taken out of the centrifuge and immediately plaged in the work area prepared by the instructor under a fume hood.
5. A fresh pair of gloves were put on as to maintain sterility and to protect the hands from harmful chemicals.
6. The designated work area was covered with bench paper to create a clean work surface and the area that the procedure took place on was an agar plate cover bottom side up with a piece of parafilm in the center and a PELCO Tab on the center of the parafilm.
7. Using EM forceps, a fresh grid was removed from a box of unused grids, touching only the very edge of the grid to not damage or contaminate it.
8. The grid was placed on the very edge of the PELCO tab, dark-and-shiny side UP.
9. Using a micropipettor, 10μl of lysate was placed onto the grid, without physically touching the grid with the pipette tip.
10. The phage was allowed to settle onto the grid for at least 2 minutes as the titer was calculated at above 109 .
11. Using a small wege of filter paper, the excess fluid was wicked off without toughing the grid.
12. Using a micropipette, 10μl of sterile water was placed onto the grid, allowed to sit for 2 minutes, and was wicked off using a fresh wedge of filter paper.
13. Then 10μl of 1% uranyl acetate was placed onto the grid, without physically touching the grid with the pipette tip.
IMPORTANT: Uranyl acetate is a very toxic compound, gloves were worn throughout this procedure to not touch the compound.
14. The 10μl of 1% Uranyl acetate was let to sit for 2 minutes, and was wicked off by using a wedge of filter paper. The UA stain was wicked multiple times until the surface of the grid was completely dry and looked like a rainbow oil slick.
15. Once the grid was completely dry, the grid was placed into a grid box for storage. (NOTE: the grid box was placed inside a vacuumed jar, so the vacuum was removed, the jar was unsealed opened, the box taken out to place the grid, and was placed back into the jar and the jar was resealed using a vacuumed.)
Results:
Once the grid was dried and had a rainbow slick color, the grid was placed inside the storage container in section A5.
Following a few days, the TEM showed the phage, which shows the morphology of a siphovirus as shown in the picture below:
Conclusion and Next Steps:
After completing the steps and placing the grid into the storage container, next will be to extract the phage DNA.
