Discovery of Agnetha
Agnetha Information
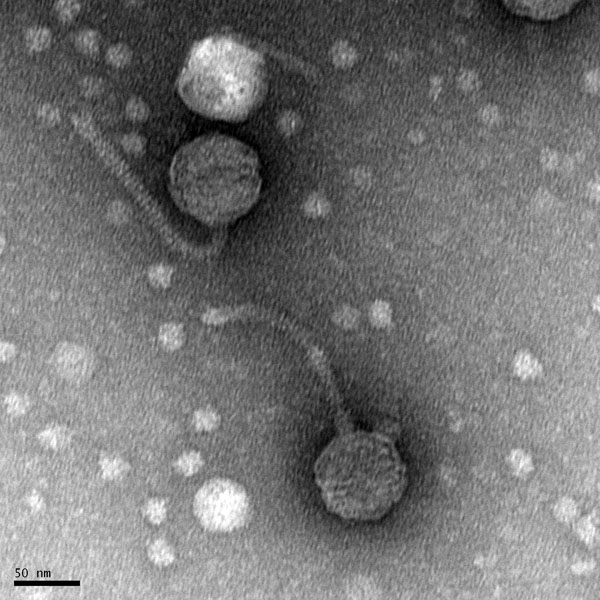
Morphology: Siphoviridae
Sample Collection
| Collector Name |
Selina Alvarado | Selina Alvarado | Selina Alvarado | Selina Alvarado | Selina Alvarado |
| Sample No. | 1 | 2 | 3 | 4 | 5 |
| Date of Collection | 08/25/2020 | 08/25/2020 | 08/26/2020 | 08/27/2020 | 08/28/2020 |
| Sample Type | Water | Water | Water | Soil (Sheep Feces) | Moist Soil |
| General Location | Forest | Forest | Barn | Barn | Garden |
| Location Description | Pond | Pond | Sheep Water Trough | Barn | Garden |
| GPS Coordinates | 32.2315763, -98.3223622 | 32.2315763, -98.3223622 | 32.2351009, -98.3182829 | 32.2351009, -98.3182829 | 32.2342890, -98.3174011 |
| Sample Depth | Surface of Water | Surface of Water | Surface of Water | Surface Level | Surface Level |
| Ambient Temperature | 30°C | 30°C | 29°C | 28°C | 33°C |
Isolation/Purification
aTitle: Direct Isolation
Date: 08/26/2020 Redo: No Sample: number 1.
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
Because this was the first isolation, some trouble was encountered. There was some difficulty remembering to use aseptic technique and stay near the bunsen burner flame to stay in the designated sterile area. This was a direct isolation with a water sample, therefore no liquid media was used.
- The work area was prepared for aseptic work.
- 2ml of sample were removed with a syringe and filtered using a 0.22ul syringe filter into a microcentrifuge tube.
- 500ul of the filtered sample were pipetted into a tube with 250ul of hot bacteria with a 100ul micropipette (100ul 5 times).
- The inoculated bacteria was left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into the inoculated bacteria tube and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plate was left undisturbed for 20 minutes.
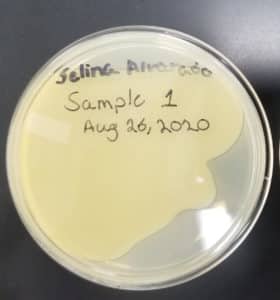
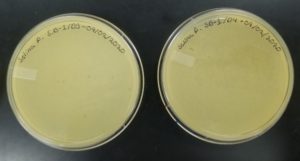
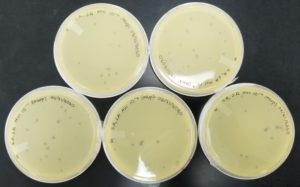
- Sample 1 plate was inverted and incubated at 11:36am at 23-30 degrees celsius.
- The plate was left in the incubator for 48 hours before removal.
Note: Unfortunately the agar plate was not tilted in time, therefore the top agar did not evenly coat the plate.
Results:
Negative result from first sample: 08/28/2020
Poor plating can clearly be seen because the top agar is not evenly spread. There was no bacteria or plaques.
Conclusions and Next Steps:
Because this was the first isolation, mishandling of equipment may have disturbed the results. This sample could not be used for the next step because no plaques appeared.
Title: Direct Isolation
Date: 08/26/2020 Redo: No Sample: number 2
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
Sample 1 and 2 were done the same day.
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 2ml of sample were removed with a syringe and filtered using a 0.22ul syringe filter into a microcentrifuge tube.
- 500ul of the filtered sample were pipetted into a tube with 250ul of hot bacteria with a 100ul micropipette (100ul 5 times).
- The inoculated bacteria was left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into the inoculated bacteria tube and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plate was left undisturbed for 20 minutes.
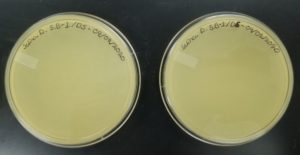
- Sample 1 plate was inverted and incubated at 11:36am at 23-30 degrees celsius.
- The plate was left in the incubator for 48 hours before removal.
Results:
The results for sample 2 were negative: 08/28/2020
Conclusions and Next Steps:
Once again the results were negative, there were no plaques to use for the next step. This sample was parafilmed and stored.
Title: Direct Isolation
Date: 08/27/2020 Redo: No Sample: number 3.
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 2ml of sample were removed with a syringe and filtered using a 0.22ul syringe filter into a microcentrifuge tube.
- 500ul of the filtered sample were pipetted into a tube with 250ul of hot bacteria with a 100ul micropipette (100ul 5 times).
- The inoculated bacteria was left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into the inoculated bacteria tube and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plate was left undisturbed for 20 minutes.
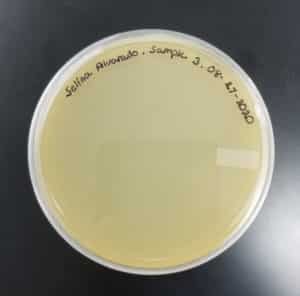
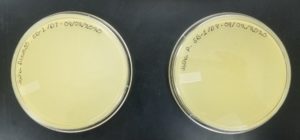
- Sample 3 plate was inverted and incubated at 4:51pm at 23-30 degrees celsius.
- The plate was left in the incubator for 48 hours before removal.
Results:
Sample 3 had negative results: 08/29/2020
Conclusion and Next Steps:
No plaques appeared, therefore sample 3 was parafilmed and stored, it could ot be used for the next step.
Title: Direct Isolation
Date: 08/28/2020 Redo: No Sample: number 4.
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
This was the first soil sample. Some trouble was encountered due to the components in the soil.
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 5ml of liquid media were pipetted into the tube with the sample and mixed thoroughly.
- The sample was placed in a shaking incubator and was left there for an hours and 15 minutes.
- The sample was distributed evently between 6 microcentrifuge tubes with a 100ul micropipette.
- The 6 microcentrifuge tubes were placed in the microcentrifuge for about 5 minutes.
- About 2ml of sample were pipetted into the syringe tube with the filter already attached.
- The sample was filtered carefully into another labeled microcentrifuge tube.
- 500ul of the filtered sample were pipetted into the tube with the host bacteria.
- The inoculated bacteria was left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into the inoculated bacteria tube and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plate was left undisturbed for 20 minutes.
- Sample 4 plate was inverted and incubated at 4:50pm at 23-30 degrees celsius.
- The plate was left in the incubator for 48 hours before removal.
Results:
Some positive results: 08/31/2020
The plate was contaminated though some small plaques were visible.
Conclusion and Next Steps:
Other bacteria could have gotten into the sample. The filtration process was longer and there was a lot of handling. This sample was parafilmed and stored. It was a candidate for the nexts steps.
Title: Direct Isolation
Date: 08/28/2020 Redo: No Sample: number 5.
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
This was also a soil sample, and it was done along with sample 4. There were no complications with the filtration process.
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 5ml of liquid media were pipetted into the tube with the sample and mixed thoroughly.
- The sample was placed in a shaking incubator and was left there for an hours and 15 minutes.
- 2ml of sample were removed with a syringe and the filter was attached.
- The sample was filtered carefully into a labeled microcentrifuge tube.
- 500ul of the filtered sample were pipetted into the tube with the host bacteria.
- The inoculated bacteria was left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into the inoculated bacteria tube and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plate was left undisturbed for 20 minutes.
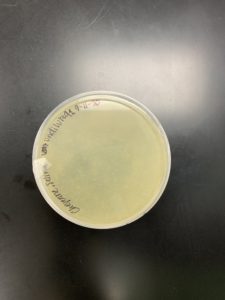
- Sample 5 plate was inverted and incubated at 4:50pm at 23-30 degrees celsius.
- The plate was left in the incubator for 48 hours before removal.
Results:
Positive results: 08/31/2020
This was the best plate out of the five. Multiple plaques were visible.
Conclusion and Next Steps:
This sample was chosen for the next step: purification. The plaques were of great quality, and no errors in plating occured.
Title: Purification (Serial Dilutions)
Date: 08/31/2020 Redo: No Sample: number 5.
Purpose: To prepare samples of decreasing concentrations with sample number five using serial dilutions and plaque assay.
Notes:
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 100ul of phage buffer were pipetted into a labeled (5.B) microcentrifuge tube.
- A plaque was carefully “picked” with a sterile 100ul micropipette tip.
- The sterile tip with the plaque was tipped/rubbed agaisnt the wall of the microcentrifuge tube (5.B) so the plaque would dislodged.
- Eight microcentrifuge tubes were aquired and labled.
- 90ul of phage buffer were pipetted into each tube using a micropipette.
- 10ul of the origional sample were pipetted into the microcentrifuge tube labled 10^1 and was vortexed.
- 10ul of tube 10^1 were pipetted into tube 10^2 and vortexed.
- 10ul of tube 10^2were pipetted into tube 10^3 and vortexed.
- 10ul of tube 10^3 were pipetted into tube 10^4 and vortexed.
- 10ul of tube 10^4 were pipetted into tube 10^5 and vortexed.
- 10ul of tube 10^5 were pipetted into tube 10^6 and vortexed.
- 10ul of tube 10^6 were pipetted into tube 10^7 and vortexed.
- 10ul of tube 10^7 were pipetted into tube 10^8 and vortexed.
- 10ul of each dilution were pipetted into labeled tubes with host bacteria. (8 tubes of host bacteria; one for each dilution).
- The tubes with inoculated bacteria were left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into each of the inoculated bacteria tubes and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plates were left undisturbed for 20 minutes.
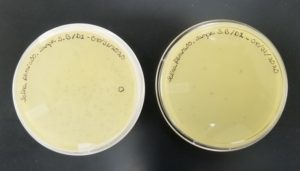
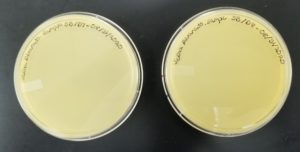
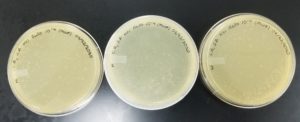
- The plates were inverted and incubated at 6:21pm at 23-30 degrees celsius.
- The plates were left in the incubator for 48 hours before removal.
Results:
Positive results: 09/02/2020
The only plates that had plaques were plates number 1 and 2. The other 6 did not have anything.
Conclusion and Next Steps:
The reason that so little plates had plaques could be because a higher concentration was needed for the dilutions. Plate number 1 was chosen to proceed to the second set of dilutions.
Title: Purification (Serial Dilutions)
Date: 09/02/2020 Redo: No Sample: number 5.B.
Purpose: To prepare samples of decreasing concentrations with sample number 5.B using serial dilutions and plaque assay.
Notes:
This is the second set of serial dilutions. The circled plaque from plate number one was used for this procedure.
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- 100ul of phage buffer were pipetted into a labeled (5.B) microcentrifuge tube.
- A plaque was carefully “picked” with a sterile 100ul micropipette tip.
- The sterile tip with the plaque was tipped/rubbed agaisnt the wall of the microcentrifuge tube (5.B-1) so the plaque would dislodged.
- Eight microcentrifuge tubes were aquired and labled.
- 90ul of phage buffer were pipetted into each tube using a micropipette.
- 10ul of sample (5.B-1) were pipetted into the microcentrifuge tube labled 10^1 and was vortexed.
- 10ul of tube 10^1 were pipetted into tube 10^2 and vortexed.
- 10ul of tube 10^2were pipetted into tube 10^3 and vortexed.
- 10ul of tube 10^3 were pipetted into tube 10^4 and vortexed.
- 10ul of tube 10^4 were pipetted into tube 10^5 and vortexed.
- 10ul of tube 10^5 were pipetted into tube 10^6 and vortexed.
- 10ul of tube 10^6 were pipetted into tube 10^7 and vortexed.
- 10ul of tube 10^7 were pipetted into tube 10^8 and vortexed.
- 10ul of each dilution were pipetted into labeled tubes with host bacteria. (8 tubes of host bacteria; one for each dilution).
- The tubes with inoculated bacteria were left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into each of the inoculated bacteria tubes and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
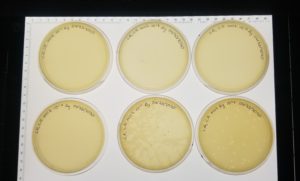
- Agar plates were left undisturbed for 20 minutes.
- The plates were not inverted (because of top agar) and incubated at 11:40am at 23-30 degrees celsius.
- The plates were left in the incubator for 48 hours before removal.
Note: Unfortunately some trouble was encountered with the top agar dude to a complication with the incubator it was in.
Results:
Negative Results: 09/02/2020
Bad plating technique is obvious. Plate number one did not solidify well and in most plates the top agar is broken.
Conclusion and Next Steps:
The reason the top agar did not solidify right was because it had to be microwaved prior to the procedure because it had solidified. After it was microwaved and it was ready to used, it was not mixed well. This caused it to be watery. This procedure had to be redone.
Title: Purification (Serial Dilutions)
Date: 09/03/2020 Redo: Yes Sample: number 5.B-1.
Purpose: To prepare good quality agar plates from the second serial dilution using plaque assay.
Notes:
This is the procedure for the redone agar plates for the second serial dilutions.
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- Eight microcentrifuge tubes were aquired and labled.
- 90ul of phage buffer were pipetted into each tube using a micropipette.
- 10ul of sample 5.B-1 were pipetted into the microcentrifuge tube labled 10^1 and was vortexed.
- 10ul of tube 10^1 were pipetted into tube 10^2 and vortexed.
- 10ul of tube 10^2were pipetted into tube 10^3 and vortexed.
- 10ul of tube 10^3 were pipetted into tube 10^4 and vortexed.
- 10ul of tube 10^4 were pipetted into tube 10^5 and vortexed.
- 10ul of tube 10^5 were pipetted into tube 10^6 and vortexed.
- 10ul of tube 10^6 were pipetted into tube 10^7 and vortexed.
- 10ul of tube 10^7 were pipetted into tube 10^8 and vortexed.
- 10ul of each dilution were pipetted into labeled tubes with host bacteria. (8 tubes of host bacteria; one for each dilution).
- The tubes with inoculated bacteria were left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipetted into each of the inoculated bacteria tubes and quicky pipetted back up.
- Top agar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plates were left undisturbed for 20 minutes.
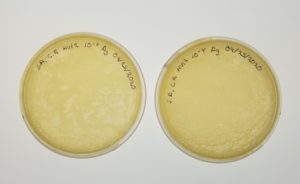
- The plates were inverted and incubated at 4.37pm at 23-30 degrees celsius.
Results:
Negative results: 09/08/2020
Some plates had plaques but no webbed plates.
Conclusion and Next Steps:
Since there were no webbed plates, the dilution ratio will be incresed to a 1:5.
Title: Purification (Serial Dilutions)
Date: 09/09/2020 Redo: Yes Sample: AG-1
Purpose: To prepare good quality agar plates from the second serial dilution using plaque assay.
Notes:
This is the procedure to attempt to get a webbed plate with an increased concentration of 1:5
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol
- Three microcentrifuge tubes were acquired and labeled.
- 160ul of phage buffer were pipetted into each tube using a micropipette.
- 40ul of tube 10^1 were pipetted into tube 10^1and vortexed.
- 40ul of tube 10^2 were pipetted into tube 10^2 and vortexed.
- 40ul of tube 10^3 were pipetted into tube 10^3 and vortexed.
- The tubes wth inoculated bacteria were left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipet Ted into each of the inoculated bacteria tubes and quickly pipetted back up.
- Top sugar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plates were left undisturbed for 20 minutes.
- The plates were inverted and and incubated at 5:30pm at 23-30 degrees Celsius.
Results:
Negative Results: 09/11/2020
Conclusion and Next Steps:
Since there were no webbed plates, the decision was made to do two plates of the undiluted phage in the hopes of getting a webbed plate.
Title: Purification (Serial Dilutions)
Date: 09/11/2020 Redo: Yes Sample: AG-1
Purpose: To prepare good quality agar plates from an undiluted sample using plaque assay.
Notes:
An undiluted sample was made by added 90uL of buffer and 10uL of sample in a microcentrifuge tube and vortexed.
- The tubes wth inoculated bacteria were left to sit for 10 minutes to allow for attachment.
- 3ml of top agar were pipet Ted into each of the inoculated bacteria tubes and quickly pipetted back up.
- Top sugar and inoculated bacteria were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plates were left undisturbed for 20 minutes.
- The plates were inverted and and incubated at 11:00 am at 23-30 degrees Celsius.
Results: Positive 09/14/2020
Conclusion and Next Steps:
Since we have webbed plates we can continue to the amplification step.
Amplification
Title: Amplification
Date: 09/14/2020 Redo: No Sample: AG-1
Purpose: With the achievement of webbed plates the goal is to begin a low volume lysate which consists of flooding a plate with a high concentration of phage and a combination of phage buffer, this allows for harvesting of the bacteria for optimal DNA extractions.
Notes:
- Work bench was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- Picked one of the webbed plates for lysate collection.
- Applied 8 mL of phage buffer to the webbed plate.
- Let plate sit for 2 hours at room temperature.
- Swirl the phage buffer.
- When incubation is complete, tilt the plate to one side, allowing the lysate to pool to one side.
- Get a .22um filter and a 5mL syringe.
- Use the syringe to aspirate the lysate from the plate then attach the filter to get rid of impurities.
- Store lysate at 4°C.
Results:
Positive Results: 09/14/2020
Conclusion and Next Steps:
Since we were able to obtain a low volume lysate from the webbed plated (shown above) we were able to get our low volume lysate. The next step is to plate in order to get a viable webbed plate for our high volume lysate.
Title: Amplification
Date:09/15/2020 Redo:No Sample: AG-1
Purpose: Plate the low volume lysate in order to get a viable plate for a high volume lysate using plaque assay.
Notes:
- Work area was prepared for aseptic work using CiDecon and Ethyl Alcohol.
- Eight microcentrifuge tubes were acquired and labeled.
- 90 ul of phage buffer were pipetted into each tube using a micropipitte.
- 10 ul of the lysate sample were pipetted into the tube labeled 10^1 and was vortexed.
- 10 ul of tube 10^1 was pipetted into tube 10^2 and vortexed.
- 10 ul of tube 20^2 was pipetted into tube 10^3 and vortexed.
- 10 ul of tube 10^3 was pipetted into tube 10^4 and vortexed.
- 10 ul of tube 10^4 was pipetted into tube 10^5 and vortexed.
- 10 ul to tube 10^5 was pipetted into tube 10^6 and vortexed.
- 10ul of tube 10^6 was pipetted into tube 10^7 and vortexed.
- 10 ul of tube 10^7 was pipetted into tube 10^8 and vortexed.
- 10 ul of each dilution were pipetted into labeled tubes with host bacteria. (8 tubes of host bacteria; one for each dilution.)
- the tubes with inoculated bacteria were left to sit for 7 minutes to allow for attachment.
- 3ml of top agar were pipetted into each of the inoculated bacteria tubes and quickly pipetted back up.
- ‘top agar and inoculated were pipetted onto an agar plate, bubbles (if any) were quickly removed.
- Agar plates were left undisturbed for 20 minutes.
- The plates were inverted and incubated at 5:30pm at 23-30 degrees celcius.
Results:
Postive Results: 09/17/2020
Conclusions and Next Steps:
The dilution of 10^4 was our best webbed plate. The next step will be to obtain our high volume lysate from it and also make multiple plates of the 10^4 to get a higher volume for future steps.
Title: Amplification
Date: 09/17/2020 Redo: No Sample: AG lvl
Purpose: To obtain a high volume lysate by flooding plates number 1, 6, 8, and 9.
Notes:
- Plates number 1, 6, 8, and 9 were flooded with 6ml of phage buffer each.
- They were allowed to sit for 2 hours.
- The phage buffer was removed and filtered using a 0.22 ul fiter and syringe.
- Serial dilutions were conducted using the high volume lysate.
- The dilutions were used to inoculate bacteria and plated following plaque assay procedure.
Results:
Postive results: 09/19/2020
Conclusions and Nest Steps:
The dilutions were succesfull and plate 10^-4 was the best out of all. This plate was chosen to produce more. The titer was not high enough, with the value of the titer being 2.1 x 10^9, more webbed plates were needed.
Title: Amplification
Date: 09/21/2020 Redo: No Sample: AG Hvl
Purpose: To obtain more webbed plates using serial dilution number 10^-4
Notes:
- The work area was prepared for aseptic work.
- Dilution number 10^-4 was used to plate 5 plates using plaque assay.
- These plates were incubated at 10:10 am and were left in the incubator for 2 days.
Results:
Negative results: 09/23/2020
Conclusion and Next Steps:
There were no webbed plates. To proceed the dilution was redo.
Title: Amplification
Date: 09/23/2020 Redo: Yes Sample: AG Hvl
Purpose: To obtain more webbed plates using serial dilution number 10^-4.
Notes:
- The working area was prepared for aseptic work.
- Dilutions 10^-1 to 10^-4 were redon using the serial dilutions procedure.
- Dilution number 10^-4 was used to inoculate bacteria and plate 3 plates using plaque assay. Each place was labled 1-3 for identification.
- Once the plates were ready, they were placed in the incubator at 10:05 am.
- Plate number 2 was removed at 12:05 pm on 09/24/2020
- Plates 1 and 3 were removed at 2:00 pm on 09/25/2020
Results:
Positive results: 09/25/2020
Conclusion and Next Steps:
All of the plates were webbed therefore we could proceed to collect more lysate to have a higher titer.
Title: Amplification
Date: 09/25/2020 Redo: No Sample: AG Hvl
Purpose: To collect more high volume lysate for a higher titer using plaque assay.
Notes:
- The work area was prepared for aseptic work.
- Dilution 10^4 plates 1-3 were flooded with 6ml of phage buffer and were left undisturbed for 2 hours.
- A syringe was used to remove the buffer with the phage from the plates. The syringe was attached to a 0.22ul filter to filter the lysate into a conical tube (HVL2).
- The new high volume lysate was used to conduct serial dilutions.
- Once diluted from 10^-1 to 10^-8, plaque assay was conducted.
- The plates were placed in the incubator at 5:40 pm and removed at 1:30 pm on 09/27/2020
Notes: Some complications were encountered with top agar on plates with the dilutions 10^-5, 10^7, and 10^-8.
Results:
Positive results: 09/28/2020
Conclusion and Next Steps:
The plates were good considering the complications witht he top agar. The titer was calculated and resulted being 6.0 x 10^9, a great value! We will proceed by archieving our bacteriophage Agnetha.
DNA Extraction
Title: DNA Extraction
Date: 10/06/2020 Redo: No
Purpose: To extract DNA from phage.
Notes:
- 5mL of high volume lysate was mixed with 20uL of nuclease in a conical tube and inverted.
- The tube was incubated at 37C for 10 minutes. When it was removed from the incubator, 1mL of the solution was pipetted into 5 microcenterfuge tubes each.
- 20mL of ZnCl2 were added to each tube and gently inverted then incubated for 5 minutes at 37C.
- The 5 tubes were centerfuged for 1 minute at 10,000 rpm.
- After the tubes were centerfuged the supernantant was removed carefully and 500uL of TES buffer were added to each tube. The tubes were incubated for 15 minutes at 60C.
- 1uL of proteinase K was added to each tube and incubated at 37C for 10 minutes.
- 60uL of potassium acetate was added to each tube and put in ice for 15 minutes.
- After the tubes were centerfuged at 4C for 1 minute at 12,000 rpm. The supernatant was removed and kept in other 5 microcenterfuge tubes and the pelletes were discarded.
- 50uL of isopropenol were added to each tube and left on ice overnight.
- The next day the tubes were centerfuged at top speed for 10 minutes and the supernatant was discarded.
- 250uL of 70% ethanol were added to each tube and spinned for 1 minute at top speed, the supernatant was discarded.
- The pelletes were dried by putting the tubes in a hot block at 38C.
- 50uL of nuclease free water was added to one tube then that solution was resuspended to the next pellet until all the tubes had been resuspended.
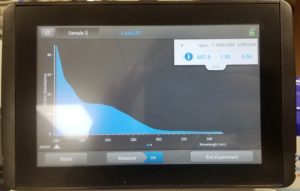
- The nanodrop reader was used to view the DNA concentration and quality.
Results:
Poor results: 10/06/2020
Conclusions and Next Steps:
Unfortunalty our numbers were not great. This was probably due to the high amount of salt in our solution. This procedure will be done again.
Title: Direct Isolation
Date: October 15, 2020 Redo: Yes
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
- 5mL of high volume lysate was mixed with 20uL of nuclease in a conical tube and inverted.
- The tube was incubated at 37C for 10 minutes. When it was removed from the incubator, 1mL of the solution was pipetted into 5 microcenterfuge tubes each.
- 20mL of ZnCl2 were added to each tube and gently inverted then incubated for 5 minutes at 37C.
- The 5 tubes were centerfuged for 1 minute at 10,000 rpm.
- After the tubes were centerfuged the supernantant was removed carefully and 500uL of TES buffer were added to each tube. The tubes were incubated for 15 minutes at 60C.
- 1uL of proteinase K was added to each tube and incubated at 37C for 10 minutes.
- 60uL of potassium acetate was added to each tube and put in ice for 15 minutes.
- After the tubes were centerfuged at 4C for 1 minute at 12,000 rpm. The supernatant was removed and kept in other 5 microcenterfuge tubes and the pelletes were discarded.
- 50uL of isopropenol were added to each tube and left on ice overnight.
- The next day the tubes were centerfuged at top speed for 10 minutes and the supernatant was discarded.
- 250uL of 70% ethanol were added to each tube and spinned for 1 minute at top speed, the supernatant was discarded.
- The pelletes were dried by putting the tubes in a hot block at 38C.
- 50uL of nuclease free water was added to one tube then that solution was resuspended to the next pellet until all the tubes had been resuspended.
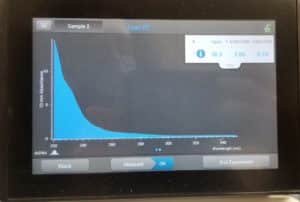
- The nanodrop reader was used to view the DNA concentration and quality.
Results:
Poor Results
Conclusions and Next Steps:
Our next steps are to obtain more high volume lysate so we can redo this procedure again in the hopes of getting better results.
Title: Direct Isolation
Date: 10/21/2020 Redo: Yes
Purpose: To extract DNA from phage.
Notes:
- 5mL of high volume lysate was mixed with 20uL of nuclease in a conical tube and inverted.
- The tube was incubated at 37C for 10 minutes. When it was removed from the incubator, 1mL of the solution was pipetted into 5 microcenterfuge tubes each.
- 20mL of ZnCl2 were added to each tube and gently inverted then incubated for 5 minutes at 37C.
- The 5 tubes were centerfuged for 1 minute at 10,000 rpm.
- After the tubes were centerfuged the supernantant was removed carefully and 500uL of TES buffer were added to each tube. The tubes were incubated for 15 minutes at 60C.
- 1uL of proteinase K was added to each tube and incubated at 37C for 10 minutes.
- 60uL of potassium acetate was added to each tube and put in ice for 15 minutes.
- One thing that changed for this time is that we were unaware that the Protenase K was supposed to have a gel-like consistency and be very white in order for it to be done. This was fixed for this time and all the remaining times we have done this.
- After the tubes were centerfuged at 4C for 1 minute at 12,000 rpm. The supernatant was removed and kept in other 5 microcenterfuge tubes and the pelletes were discarded.
- 50uL of isopropenol were added to each tube and left on ice overnight.
- The next day on Monday the 26th the tubes were centerfuged at top speed for 10 minutes and the supernatant was discarded.
- 250uL of 70% ethanol were added to each tube and spinned for 1 minute at top speed, the supernatant was discarded.
- The pelletes were dried by putting the tubes in a hot block at 38C.
- 50uL of nuclease free water was added to one tube then that solution was resuspended to the next pellet until all the tubes had been resuspended.
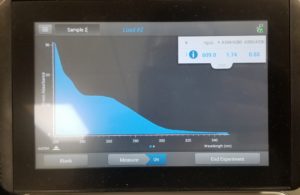
- The nanodrop reader was used to view the DNA concentration and quality.
Results:
Poor results: 10/28/2020
Conclusion and Next Steps:
Since our numbers were not great this procedure is going to be done again.
Title: Direct Isolation
Date:10/28/2020 Redo: Yes
Purpose: To extract DNA from phage.
Notes:
- 5mL of high volume lysate was mixed with 20uL of nuclease in a conical tube and inverted.
- The tube was incubated at 37C for 10 minutes. When it was removed from the incubator, 1mL of the solution was pipetted into 5 microcenterfuge tubes each.
- 20mL of ZnCl2 were added to each tube and gently inverted then incubated for 5 minutes at 37C.
- The 5 tubes were centerfuged for 1 minute at 10,000 rpm.
- After the tubes were centerfuged the supernantant was removed carefully and 500uL of TES buffer were added to each tube. The tubes were incubated for 15 minutes at 60C.
- 1uL of proteinase K was added to each tube and incubated at 37C for 10 minutes.
- 60uL of potassium acetate was added to each tube and put in ice for 15 minutes.
- Waited for the gel-like consistency and for the mixture to turn very white before continuing on to the next step.
- After the tubes were centerfuged at 4C for 1 minute at 12,000 rpm. The supernatant was removed and kept in other 5 microcenterfuge tubes and the pelletes were discarded.
- The resuspension was not going according to plan, so the process was tweaked. This entailed resuspending twice so that the pellet was no longer together before continuing to the next step.
- 50uL of isopropenol were added to each tube and left on ice overnight.
- The next day on October 30th, the tubes were centerfuged at top speed for 10 minutes and the supernatant was discarded.
- 250uL of 70% ethanol were added to each tube and spinned for 1 minute at top speed, the supernatant was discarded.
- The pelletes were dried by putting the tubes in a hot block at 38C.
- 50uL of nuclease free water was added to one tube then that solution was resuspended to the next pellet until all the tubes had been resuspended.
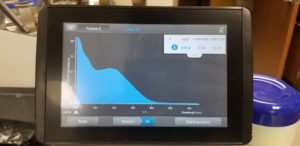
- The nanodrop reader was used to view the DNA concentration and quality.
Results:
Poor Results: 10/30/2020
Conclusion and Next Steps
Unfortunetly, the numbers were not good so this procedure is going to be repeated.
Title: Direct Isolation
Date: 11-09-2020 Redo: Yes
Purpose: To extract DNA from phage
Notes:
- 5mL of high volume lysate was mixed with 20uL of nuclease in a conical tube and inverted.
- The tube was incubated at 37C for 10 minutes. When it was removed from the incubator, 1mL of the solution was pipetted into 5 microcenterfuge tubes each.
- 20mL of ZnCl2 were added to each tube and gently inverted then incubated for 5 minutes at 37C.
- The 5 tubes were centerfuged for 1 minute at 10,000 rpm.
- After the tubes were centerfuged the supernantant was removed carefully and 500uL of TES buffer were added to each tube. The tubes were incubated for 15 minutes at 60C.
- 1uL of proteinase K was added to each tube and incubated at 37C for 10 minutes.
- 60uL of potassium acetate was added to each tube and put in ice for 15 minutes.
- Waited for the gel-like consistency and for the mixture to turn very white before continuing on to the next step.
- Had D.Edwards assist in making sure the consistency was right before continuing the next step.
- After the tubes were centerfuged at 4C for 1 minute at 12,000 rpm. The supernatant was removed and kept in other 5 microcenterfuge tubes and the pelletes were discarded.
- The resuspension was not going according to plan, so the process was tweaked. This entailed resuspending twice so that the pellet was no longer together before continuing to the next step.
- 50uL of isopropenol were added to each tube and left on ice overnight.
- The next day on November 10, the tubes were centerfuged at top speed for 10 minutes and the supernatant was discarded.
- This was slightly different from the previous attempts because the tubes were left for two days in earlier procedures, but this one was done immediately the second day.
- 250uL of 70% ethanol were added to each tube and spinned for 1 minute at top speed, the supernatant was discarded.
- The pelletes were dried by putting the tubes in a hot block at 38C.
- 50uL of nuclease free water was added to one tube then that solution was resuspended to the next pellet until all the tubes had been resuspended.
- The nanodrop reader was used to view the DNA concentration and quality.
- Also tried it with the Qubit to get the concentration.
Results:
Poor results on: 11/09/2020

Conclusion:
We are going to make some changes and then try the procedure again. We combined the same DNA from the same high volume lysate and it came out to be about 70ul, and we are going to add ethanol to it then resuspend. So it’s going to be a repeat of Day 2 without doing everything for Day 1.
Title: DNA Precipitation
Date: 11/11/2020 Redo: No
Purpose: Concentrate the DNA even more than it already is
Notes:
- We combined the same DNA from the same high volume lysate and it came out to be about 70ul, and we are going to add ethanol to it then resuspend.
- Added 7ul of 3M Na-Acetate and 175ul of ice-cold 100% ethanol to our mixed DNA sample.
- Stores at -20°C for at least 1 hour to precipitate DNA
- Centrifuge at full speed for 20 minutes.
- Let the pellets air-dry.
- Resuspend the DNA in a suitable volume of sterile TE buffer or distilled water.
Results:
Characterization
Title: Direct Isolation
Date: 09/30/2020Redo: No
Purpose: To prepare the phage sample for viewing with a transmission electron microscope.
Notes:
- Prepared for aseptic work.
- Transfer 1 ml of high-titer lysate into a microcentrifuge tube.
- Balance the tube and centrifuge for a hour at 4°C
- Using a micropipette remove as much supernatant without bothering the concentrate phage at the bottom of the tube.
- Add 100 ul of phage buffer and let resuspend at 4°C for one hour.
- Move to autoclave to begin the next portion of the experiment.
- Prepare for aseptic work.
- Cover bench with bench paper.
- Get/open a Petri dish and on the top portion place a piece of parafilm in the middle of the cover.
- Place a PELCO tab and expose the adhesive.
- Using EM forceps, remove a fresh grid from the box and stick it the side of the PELCO tab, make sure that the grid is shiny side up.
- Using a micropipettor, place 10 ul of lysate on the grid without touching the tip to the grid and allow it to sit for 2 minutes.
- Using a small wedge of filter paper wick off the excess fluid.
- Rinse the grid two times with sterile water.
- Add 10 ul of 1% of uranyl acetate to the grid, leave for 2 minutes and the wick of excess with filter paper.
- Rinse the grid two times with sterile water.
- The grid should have an rainbow sheen to it.
- Place the grid in the designated grid box.
- Transport the sample to EM facility for imaging.
Results:
Text
Conclusions and Next Steps:
Text
Title: Direct Isolation
Date: month/day/year Redo: Yes or No
Purpose: This procedure is to extract phages from the environmental sample and infect the host bacteria using a plaque assay.
Notes:
Text
Results:
Text
Conclusions and Next Steps:
Text