Discovery of Gustopher
September 5th, 2018
Soil sample 1 was collected in Hamilton,Texas on September 3rd, 2018. It was located at Latitude 31.5753 Longitude -98.1510, at 85 degrees Fahrenheit.
Direct Isolation of Soil Sample 1:
- Clean up area
- Fill the 15ml conical tube 1/3 or ½ full of soil
- Add 2-3ml of media/ make sure it is submerged
- Shake the tube and mix
- Incubate the tube for 1-2 hours at 250 rpm (put in at 3:30 and took out at 4:54pm)
- Wait for soil to sit at the bottom and compact
- Filter it through syringe into small tube
- Mix it with host bacteria and wait 5-10 minutes
- 3ml of agar in the host bacteria tube
- put it into the agar plate
-
place agar plate in the incubator at 23c at 5:41pm.
September 7th, 2018
- Direct isolation is negative for phage
Soil Sample 2 was collected in Hamilton, Texas on September 3, 2018. It was located at Latitude 31.5748 Longitude -98.1509, at 85 degrees Fahrenheit.
Enriched Isolation of Soil Sample 2:
- Clean up area
- Filled the 50 ml cronical tube with soil to the 15 ml
- Added liquid media to the 35 ml mark and vortex (shake)
- Incubated the tube for an hour and a half (put in at 3:00pm and took out at 4:27pm)
- Filtered and added media to get phage to 25 ml
- Put back into incubator at 4:50pm
ERROR: Did not seed the culture with host bacteria
September 12th, 2018
- Took sample out of incubator
- Filtered the sample using a .22um filter by removing the plunger and attach a sterile filter
- Using a pipette transferred 1.4 ml from sample into two separate micro centrifuge tubes
- Pipette 1ml of sample out of each micro centrifuge tube into the syringe
- Depressed the syringe through the filter into a new micro centrifuge tube
Spot test
- Grabbed new agar plate and defrosted to room temperature
- Separated bottom of plate into as many sections as you have samples ( 2 sections: 1 sample and 1 negative control)
- Once defrosted grabbed a tube of 250ul of host bacteria
- Took 3ml of molten top agar and placed into tube of host bacteria, draw solution back into pipette, and quickly placed on agar plate.
- Once hardened placed 10ul of each sample and a negative control (buffer) on the plate
- Put incubator and Incubated for 24-48 hours to see if a plaque forms
NOTE: When performing spot test the enriched soil sample got onto the negative control side but did not touch. Plate was checked on September 13th and 14th and was negative for phage.
September 14th, 2018
Hamilton water sample was collected in Hamilton, Texas on September 3rd, 2018. It was located at Latitude 31.5755 Longitude -98.1510, at 85 degrees Fahrenheit.
Enriched isolation of Hamilton water sample 1 (D on spot test)
- Cleaned off work space
- 5ml water, 20 ml of 1x media in a 50ml conical tube (was supposed to use 45ml of media but the rest were contaminated)
- put in incubator 9/14/18 at 1:30pm
- taken out 9/14 at 3:30pm.
- disclaimer: 10x buffer was made instead of 10x media so process was delayed
- centrifuge for 10 minutes at 9607 rpm at 26 degrees Celsius
- added 10 microliters of 10x media to host bacteria, put the host bacteria into sample and put in incubator on 9/14 at 4:50pm
September 18th, 2018
Enriched isolation of Hamilton water sample 1 continued
- Took sample out of incubator
- Filtered the sample using a .22um filter by removing the plunger and attach a sterile filter
- Using a pipette transferred 1.4 ml from sample into two separate micro centrifuge tubes
- Pipette 1ml of sample out of each micro centrifuge tube into the syringe
- Depressed the syringe through the filter into a new micro centrifuge tube
Stored in fridge at 4 degrees celsius until spot test performed on September 25th.
Enriched isolation of Hamilton water sample 2 (C on spot test)
- clean area/ gather supplies
- get two 50ml conical tubes
- 9 ml of sample, 1 ml of 10x media
- vortex
- incubate at 4:00pm at 180 rpm 26 degrees Celsius
- took out 6:11pm
- centrifuged for 10 min at 9607 rpm at 16 degrees Celsius
- Filtered using .22 ul filter
- Added 250 ul of host bacteria
- Put in incubator
Taken out of incubator on September 24th, 2018. Filtered and spot test on September 25th,2018.
Spot test (refer to September 26th under Discovery of Gustopher)
A and B- Gustopher tested positive for phage
C- Hamilton water sample 2 negative for phage
D- Hamilton water sample 1 negative for phage
NOTE: At this point Direct soil sample 1, Enriched soil sample 2, and Hamilton water samples 1 and 2 tested negative for phage so I proceeded protocol with Gustopher.
Discovery of Gustopher
Found in Stephenville, Texas on September 15th, 2018
GPS coordinates lat: 32.2119 lon: -98.2042
Water conditions Turbid, cloudy
Environmental temperature 84 degrees Fahrenheit
September 18th, 2018
Enriched Isolation of Bosque river sample
Using 10x media for water sample
- cleaned area/gathered supplies
- Collected two 50ml conical tubes
- 45ml of sample 5 ml of 10x media
- vortex
- put in incubator at 3:45pm at 180rpm and 26 degrees Celsius
- took out at 5:37pm
- centrifuged 10 min at 9607 rpm at 14 degrees Celsius
- filtered with .22 microliter filter
- added 2.7 ml of 10x media
- added 250 microliters of host bacteria
- put it in incubator at 6:08pm
September 24th, 2018
- Sample taken out of incubator around 1:00pm and stored in fridge
September 25th, 2018
Filter the enriched culture
- Took sample out of fridge around 3:30pm
- Filtered the sample using a .22um filter
- Removed the plunger and attach a sterile filter
- Using a pipette transferred 1.4 ml from sample into two separate micro centrifuge tubes
- Pipette 1ml of sample out of each micro centrifuge tube into the syringe
- Depressed the syringe through the filter into a new micro centrifuge tube
Spot test
- Grabbed new agar plate and defrosted
- Separated bottom of plate into as many sections as you have samples ( 4 samples and a negative control)
- Once defrosted grabbed a tube of 250ul of host bacteria
- Took 3ml of molten top agar and placed into tube of host bacteria, draw solution back into pipette, and quickly placed on agar plate.
- Once hardened placed 10ul of each sample and a negative control (buffer) on the plate
- Put incubator around 5:00pm and Incubated for 24-48 hours to see if a plaque forms
September 26th, 2018
- Spot test has phage (A and B are both from the Bosque river, just filtered differently)
September 27th, 2018
Serial Dilutions and Plaque Assay
Serial Dilutions
- Took filtered sample out of fridge
- Collected 10 microcentrifuge tubes
- put 90ul of buffer in each labeled micro centrifuge tube
- pipette 10ul of sample into 10-1 micro centrifuge tube and vortex
- pipette 10ul of 10-1 in 10-2 tube and vortex and proceed dilution until 10-10
Plaque Assay
- Grabbed 10 host bacteria tubes and label them 10-1 to 10-10
- Pipette 10ul of 10-1 dilution into the host bacteria tube and let sit for 15 minutes to allow attachment
- Repeated for each dilution
- Collected 10 agar plates, allow to reach to room temperature, and label each one
- Took 3ml of molten top agar and place into tube of host bacteria, draw solution back into pipette, and quickly place on agar plate.
- Spread molten top agar so the entire surface was covered and allowed to harden
- Repeated for each dilution
- Once hardened, flipped plates, and put in incubator
- Put plaque assay agar plates in incubator
October 1st, 2018
- Plaque assay contaminated
October 2nd, 2018
plaque assay
Using previous serial dilutions perform plaque assay
- Grabbed 10 host bacteria tubes and label them 10-1 to 10-10
- Pipette 10ul of 10-1 dilution into the host bacteria tube and let sit for 15 minutes to allow attachment
- Repeated for each dilution
- Collected 10 agar plates, allow to reach to room temperature, and label each one
- Took 3ml of molten top agar and place into tube of host bacteria, draw solution back into pipette, and quickly place on agar plate.
- Spread molten top agar so the entire surface is covered and allow to harden
- Repeated for each dilution
- Once hardened, flipped plates, and put in incubator
October 3rd, 2018
NOTE: Many plates were contaminated and had to remake bacteria hosts.
October 8th, 2018
serial dilutions and plaque assay
serial dilutions
- re-filtered sample from fridge with a .22ul filter and syringe
- put 90ul of buffer in each labeled micro centrifuge tube
- pipette 10ul of sample into 10-1 micro centrifuge tube and vortex
- pipette 10ul of 10-1 in 10-2 tube and vortex and proceed dilution until 10-10
plaque assay
- Grabbed 10 host bacteria tubes and label them 10-1 to 10-10
- Pipette 10ul of 10-1 dilution into the host bacteria tube and let sit for 15 minutes to allow attachment
- Repeated for each dilution
- Collected 10 agar plates, allowed to reach to room temperature, and labeled each one
- Took 3ml of molten top agar and placed into tube of host bacteria, draw solution back into pipette, and quickly placed on agar plate.
- Shake or spread molten top agar so the entire surface is covered and allow to harden
- Repeat for each dilution
- Once hardened, flipped plates, and put in incubator at 9:30pm
October 9th, 2018
- 10^-3 beginning of webbed plate
- No contamination
October 10th, 2018


- Webbed plate 10-3
- 10^-4 has gradient
- Serial dilutions are seen
October 11th, 2018
Flood webbed plates
- Pipette 8ml of phage buffer into webbed plate and put in fridge at 4C
October 16th, 2018
Harvest lysate
- Cleaned bench
- Tilted the agar plate to the side and with a syringe collect all of the lysate
- Attach to a 20 ul filter and filter the lysate into a 15ml conical tube. (Only collected 4ml of filtered lysate)
Made more plates of 10^-3 to collect more lysate
- Collected 4 agar plates and allowed to defrost to room temperature
- Collected 4 tubes of bacteria host
- Used 10^-3 from previous serial dilution and pipette 10ul into each host bacteria tube
- Allowed to attach for 10-15 minutes
- Plated agar using 3ml of molten top agar and host bacteria
- Put in incubator
October 17th, 2018
- Plates were contaminated and were not concentrated enough. No webbed plate therefore cannot collect more lysate.
October 22nd, 2018
Note: I flooded the plate before a second round of purification. In order to fix this I did serial dilutions up to 10^-3 and plated them.
Serial dilutions
- 10 ul of lysate
- 90 ul of buffer in each micro centrifuge tube labeled 10^1,10^-2, and 10^-3.
- pipette 10 ul of lysate into 10^-1 and vortex, pipette 10ul of 10^-1 into 10^-2 and vortex, pipette 10ul of 10^-2 into 10^-3 and vortex.
Plaque Assay
- Grabbed 3 host bacteria tubes
- pipette 10 ul of each dilution into according tubes
- Allowed 15-20 minutes for attachment
- Pipette 3ml of top agar in tube of host bacteria/serial dilution, pipette it back, and then place on the agar plate
- Allowed to set, flipped over and placed in incubator
October 23rd, 2018
- Didn’t work/ nothing showed up
Note: Since the bacteria didn’t grown and I accidentally threw away all of my plates from the last plaque assay I had to go back to the spot test and pick a plaque
Picking a plaque
- Plaque is clear
- Placed a pipette tip in the plaque and mixed it in a micro centrifuge tube with 90ul of buffer
- Performed serial dilutions and plaque assay
- Put in incubator
October 25th, 2018
- plaque morphologies consistent
Harvesting lysate
- Took webbed plate and flooded it with 8ml of phage buffer
- put lysate in fridge at 4:30pm
- collected 6ml of small volume lysate at 11:48pm
- Using a syringe and filter took lysate out and put through a filter into a 15ml conical tube
Full plate titer from lysate
Serial dilutions
- collected 9 microcentrifuge tubes and labeled one for lysate and the rest from 10^-1 to 10^-8
- Using 30ul of lysate put in a micro centrifuge tube
- Perform serial dilutions by pipetting 10ul of each sample into the next and vortex
Plaque assay
- Collected 8 plates and label from 10^-1 to 10^-8
- put 10ul of each dilution into a host bacteria tube
- allowed 15 min for attachment
- plated using 3ml of molten top agar and the host bacteria/dilution tube for each plate
- made 2 webbed plates from 10^-2 dilution of lysate as well in order to collect more high volume lysate
October 27, 2018
- taken out around 12:30pm
October 29, 2018
- contaminated
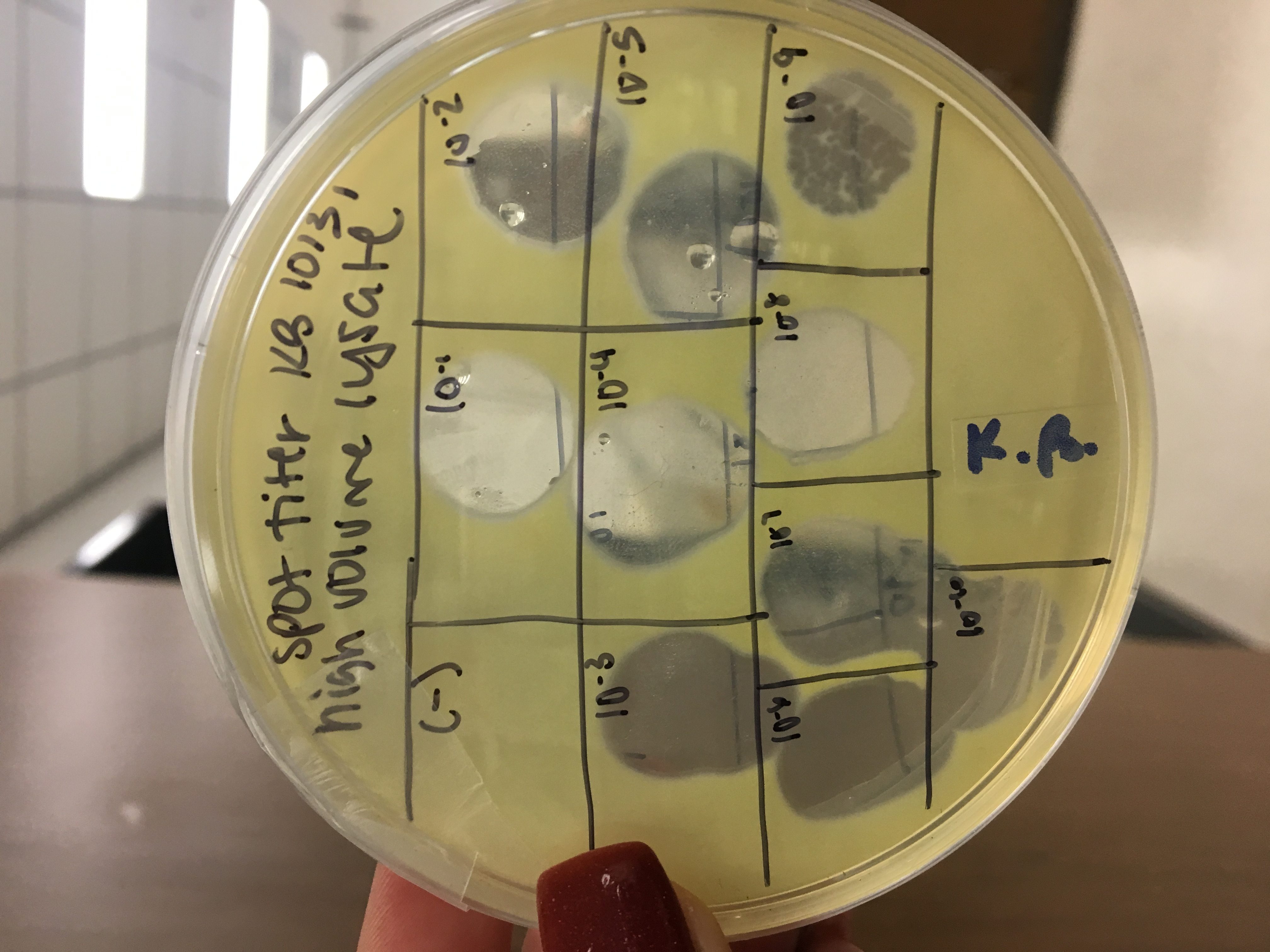
Full plate titer from small volume lysate
Serial Dilutions
- Collected 8 microcentrifuge tubes and labeled one for lysate and the rest 10^-1 to 10^-7
- Took lysate out of the fridge and put a small amount in a micro centrifuge tube
- Performed serial dilutions by pipetting 10ul of each sample into the next and vortex
plaque assay
- Collected 7 plates and labeled from 10^-1 to 10^-7
- Put 10ul of each dilution into a host bacteria tube
- allowed 15 minutes for attachment
- plated using 3ml of molten top agar and the host bacteria/dilution tube for each plate
- put in incubator at 10:30pm
October 30, 2018
Collecting high volume lysate
- Flooded plate 10^-6 at 4:00pm with 8ml of phage buffer
- Collected lysate at 8:20pm with a syringe and filter into a 15ml conical tube
- Collected about 6ml
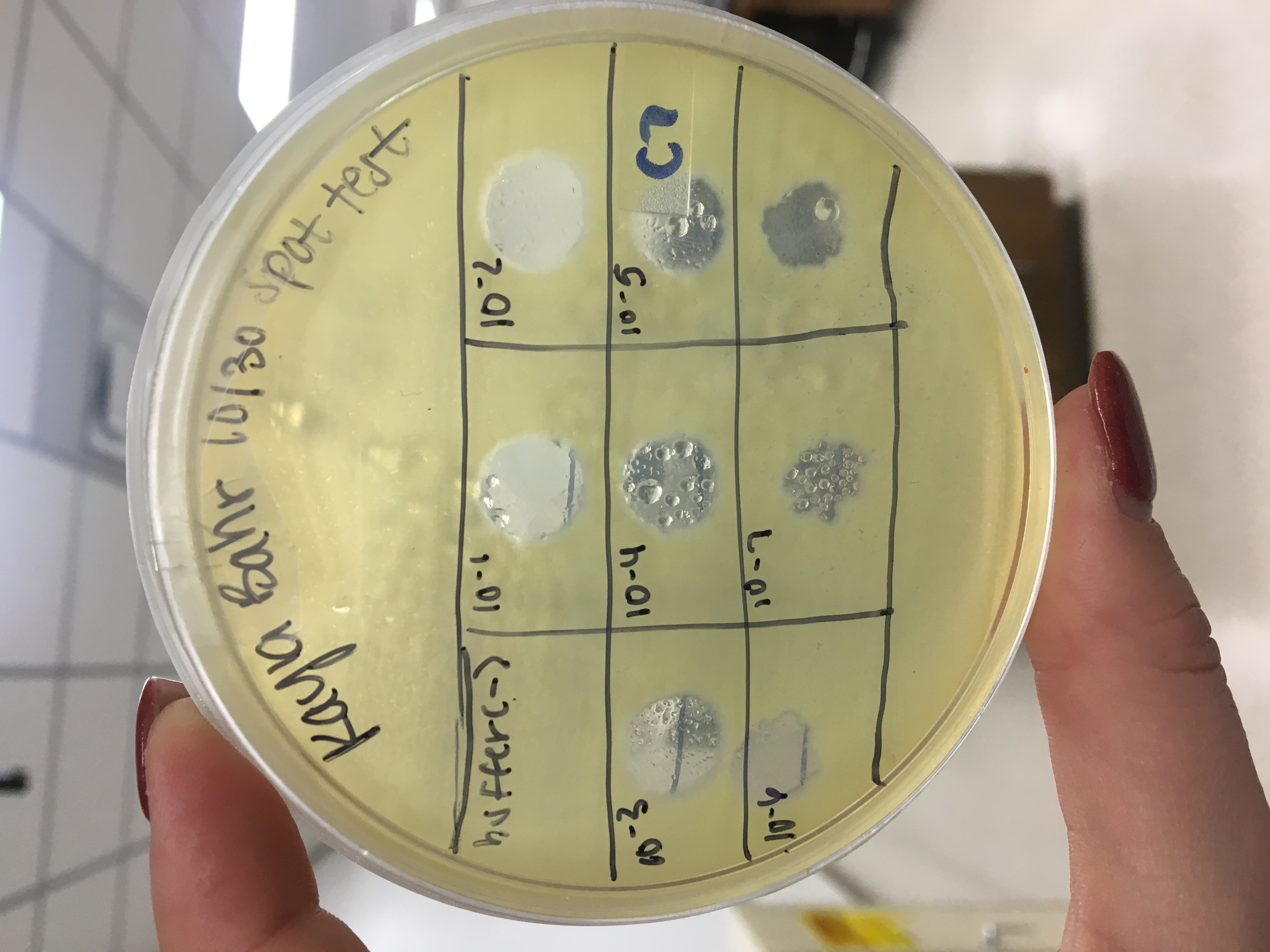
Spot titer with high volume lysate serial dilutions
- Performed spot titer in order to calculate titer because concentration is needed from high volume lysate
- Labeled an agar plate into 8 sections; one for the negative control (buffer) and 7 for the dilutions 10^-1 to 10^-7
- Grabbed dilutions from the day before from fridge and placed 3ul of each dilution/negative control into the appropriate sections
- Allowed 30 minutes to sit
- Put in incubator around 4:00pm
Plated 2 plates to 10^-6 from small volume lysate serial dilutions to make webbed plates, flood, collect more high volume lysate
Electron Microscopy
Mounting phage samples for TEM and staining using Uranyl Acetate
- put 1ml of high volume lysate in micro centrifuge tube
- balanced the centrifuge and put in for 1 hour
- small yellow pellet was present at bottom of tube
- Pipette supernatant out leaving a small amount and pellet at bottom
- put 100ul of phage buffer and put in fridge for 40 minutes
- Put a square of parafilm on a petri dish lid
- put a PELCO tab in the center and carefully placed the grid to the right of the tab with reverse tweezers
- Put 10ul of lysate without touching the tip to the grid and let sit for 2 minutes
- Wicked liquid off with filter paper
- Put 10ul of water and let sit for 2 minutes
- Wicked liquid off with filter paper
- Put 10ul of water and let sit for 2 minutes
- Wicked liquid off with filter paper
- Put 10ul of Uranyl Acetate and let sit for 2 minutes
- Wicked off liquid multiple times until looks like oil/rainbow
- let Dry and put in slot 5
- Repeated with another grid and put in slot 6
October 31st, 2018
- Flooded 2 plates and collected more high volume lysate with a total of 10ml
DNA extraction (5ml of high volume lysate)
(Insert protocol)
Too much salt so have to do again
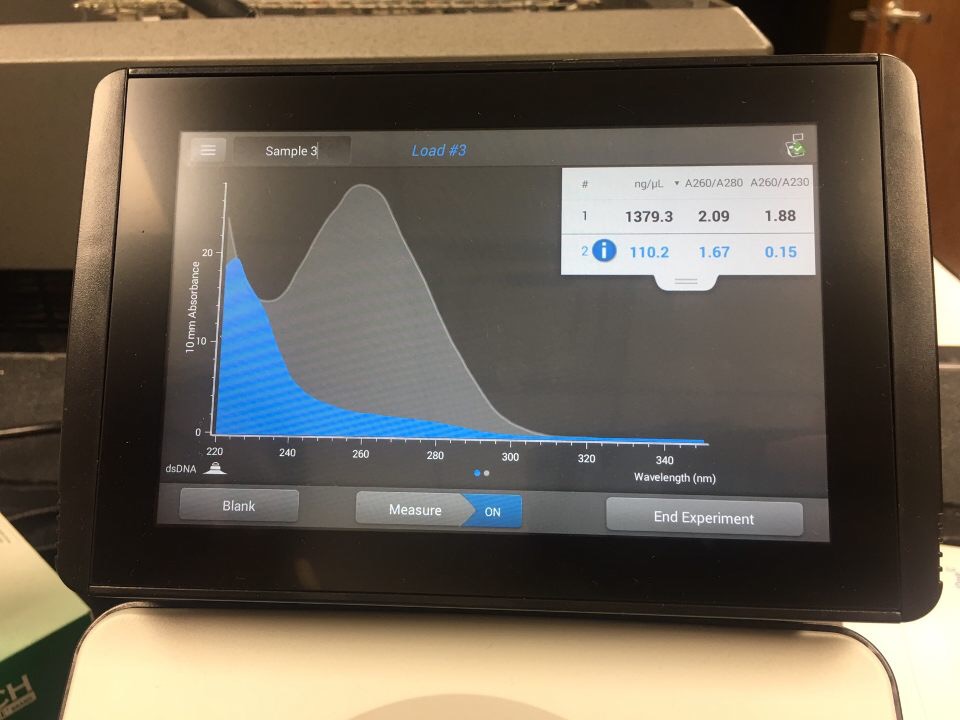
November 8th, 2018
DNA extraction (5ml of high volume lysate)
succesful
Gustopher is 1
November 17th, 2018
High volume lysate serial dilutions
spot titer
November 18th, 2018
Restriction enzyme digest/ run gel
calculate titer:
(24/3) x (10^3)(10^9)=
8 x 10^12














