Discovery of RubberBandz
September 4, 2018
- Collected a soil sample(sample 1) behind the air conditioning off the side of my brother’s work in Bluff Dale, Texas, around 10 pm at night, digging roughly 3 cm deep. It was sprinkling around 73 degrees Fahrenheit. The exact GPS location was -32.351675, -98.022275. The soil was finer, more like sand. Collected from where water leaks out of the air conditioning, so the soil is consistently damp.
September 5, 2018
- Extracted sample 1 using direct isolation technique.
- Added liquid media to a 15mL tube until the sample was submerged by about 3 mL of media. Capped the tube and inverted several times to mix until there were no noticeably large soil chunks
- Let incubate at 27 degrees Celsius in a shaking incubator for 2 hours.
- Allowed soil in sample to settle before preparing the filtrate.
- Preparing a phage filtrate
- Used a syringe to remove around 2 mL of liquid media/soil sample.
- Attached the syringe to a sterile 0.22 microliter syringe filter then dispensed around 1.5 mL of sample into a labeled 1.5 mL tube.
- Plaque Assay
- A 250 microliter aliquot of M. foliorum was obtained and gently mixed until homogenous.
- After homogenous, 500 microliters of phage sample was dispensed into the tube containing host bacteria.
- Gently mixed the host bacteria and phage sample with a gentle tilting. The tube sat on the bench for 10 minutes to allow any potential phages to attach.
- 3 mL of hot top agar was aspirated and dispensed into the host bacteria tube and then the mixed was reaspirated and plated on an agar plate. The agar plate was too cold still, allowing water to condense between the two layers of agar. The top agar flopped off the plated agar when flipped.
- After losing the samples do to flopping onto the plate lid, the above protocol was repeated using a ROOM TEMPERATURE plate. I let the plate sit for 40 minutes until Dr. Edwards came and flipped it for me because I was afraid of losing my sample again.
- Incubated plated sample for roughly between 49-50 hours.
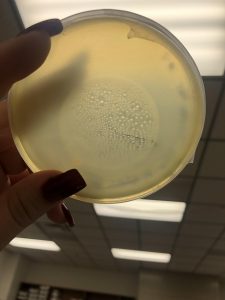
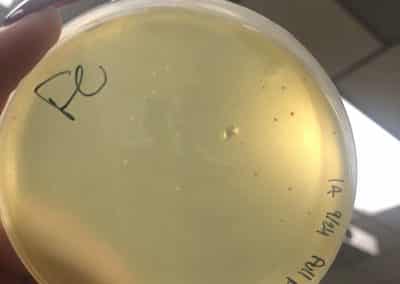
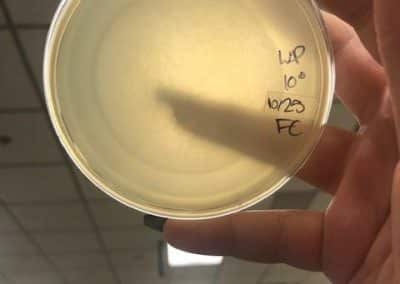
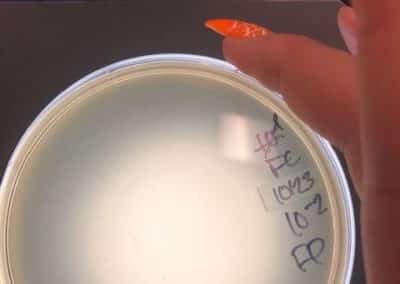
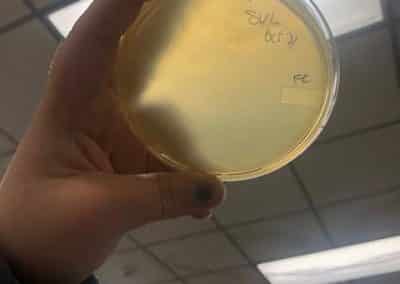
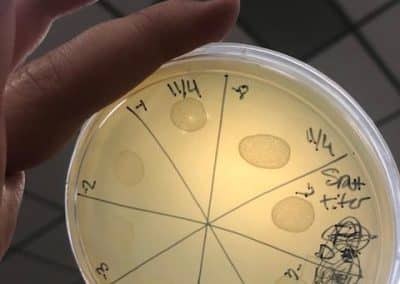
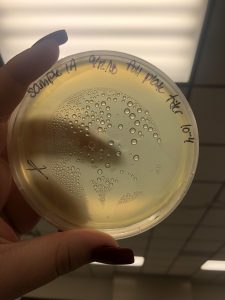
Photo taken on September 7, 2018

September 7, 2018
- Took the above photos and picked plaques 1A, 1B, and 1C.
- 1A was a small plaque with clear, turbid edges.
- 1B and 1C were also small plaques, but had unclear edges with cloudy centers.
- Actually picking the plaque
- Labeled three 1.5 mL tubes and labeled them 1A, 1B, and 1C.
- Pipetted 100 microliters of phage buffer into each tube.
- Gently stabbed plaque 1A with a sterile 200 microliter pipette tip and then swirled its respective tube and vigorously pipetted up and down to inoculate the buffer with phage.
- Discarded the pipette tip.
- Mixed sample by vortexing for a second.
- Repeated steps 3-5 for plaques 1B and 1C
- Spot test
- Gently mixed 250 microliters of the host bacteria until homogeneous and then pipetted in 3 mL of hot top agar. Aspirated all of the bacteria/agar mixed and plated it on a room temperature agar plate.
- Waited 20 minutes for the agar plate to solidify.
- Flipped plate and divided plate into three sections labeled 1A, 1B, and 1C.
- Spotted 10 microliters of each sample onto its respective labeled area.
- Let the agar absorb the spot for 15 minutes.
- Incubated the plates at 29 degrees Celsius for 59 hours.
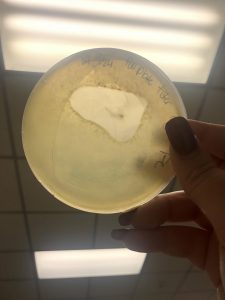
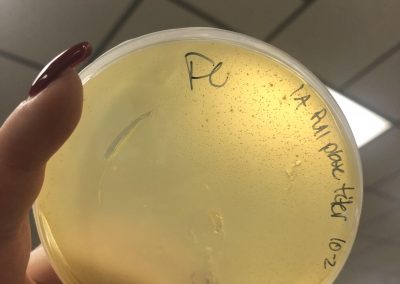
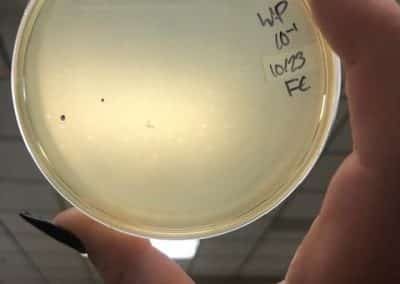
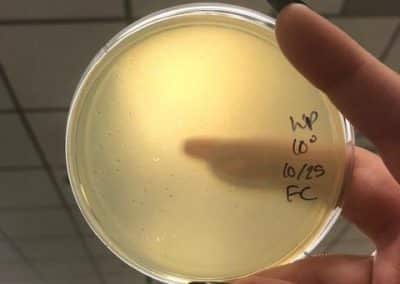
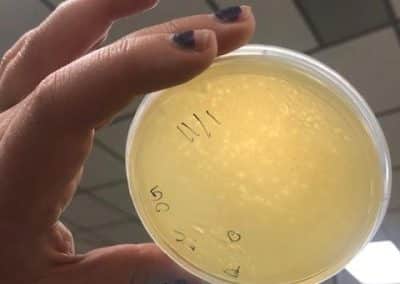
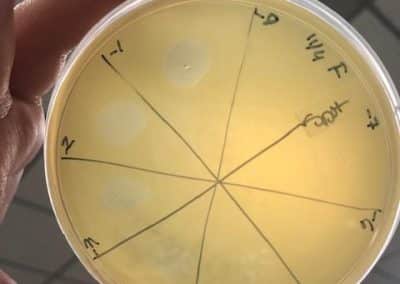
Photo taken on September 10, 2018
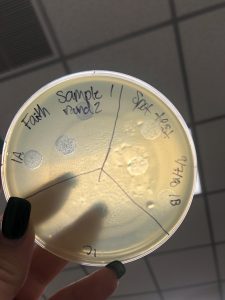
September 10, 2018
- Picked plaques from the above photo from plaques 1A and 1B.
- used the same protocol used on September 7, 2018.
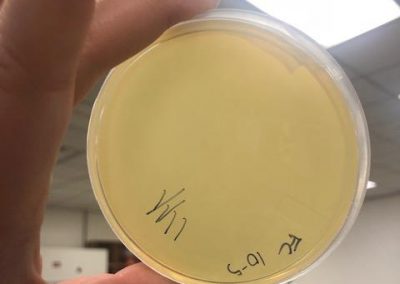
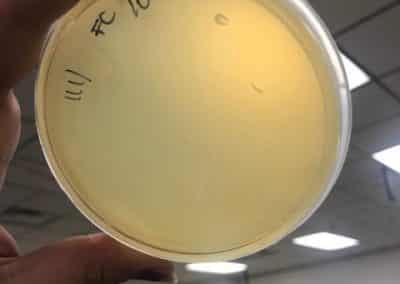
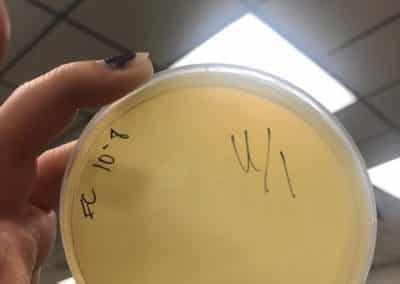
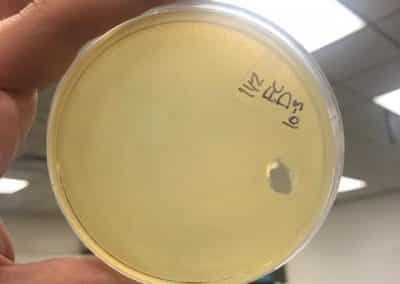
- Serial dilutions for Spot Titer
- used the picked plaque liquid from the above steps. Those were labeled OG.
- Dispensed 90 microliters of phage nine 1.5 microcentrifuge tubes labeled 10^-1, 10^-2, 10^-3, 10^-4, 10^-5, 10^-6, 10^-7, 10^-8, and 10^-9.
- 10 microliters from the OG tubes were taken and dispensed into to 10^-1 tubes. The tubes were lightly votexed to mix.
- 10 microliters from the 10^-1 tube was dispensed into the 10^-2 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-2 tube was dispensed into the 10^-3 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-3 tube was dispensed into the 10^-4 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-4 tube was dispensed into the 10^-5 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-5 tube was dispensed into the 10^-6 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-6 tube was dispensed into the 10^-7 tube and then lightly vortexed to mix.
- 10 microliters from the 10^-7 tube was dispensed into the 10^-8 tube and then lightly vortexed to mix.
- Spot Titer
- The 10 microliters of OG-10^-8 serial dilutions for plaques 1A and 1B were plated on plates prepared with a bacterial lawn like was prepared for the spot test on September 7, 2018. In total there were 10 individual spots plated that were left to absorb for 25 minutes.
- Plates were incubated at 29 degrees Celsius for 47 hours.
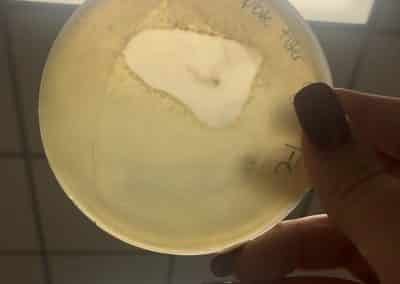
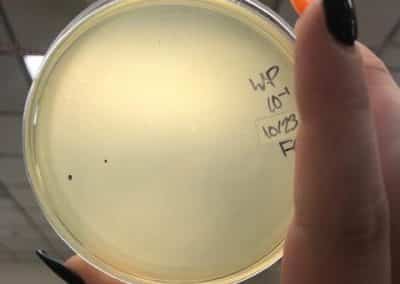
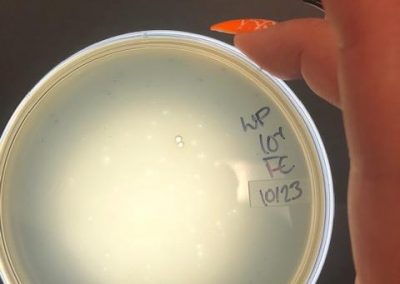
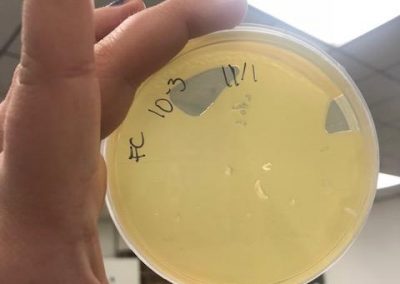
Photos from September 12, 2018


After performing a spot titer, I donated 1B to Shey and Abby and named 1A Halloweenie. 1A will be referred to as my phage sample or Halloweenie from this point in the document forward.
The spot titer for Halloweenie showed that it does infect well after the 10^-4 dilution, allowing me to only do full plate titers of the 10^-2, 10^-3, and 10^-4 samples.
September 12, 2018
- Full plate titers
- Picked a plaque from my spot titer plate and diluted it down to 10^-4 using the same procedure used on September 10, 2018 when performing spot dilutions.
- Plated dilutions 10^-2, 10^-3, and 10^-4 on bacterial lawns like those prepared on September 10, 2018.
- These plates were contaminated 🙁
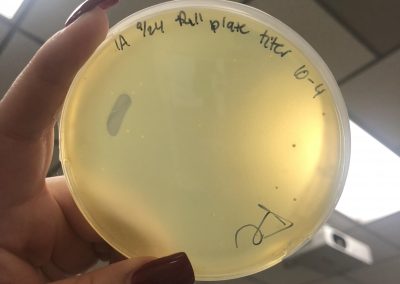
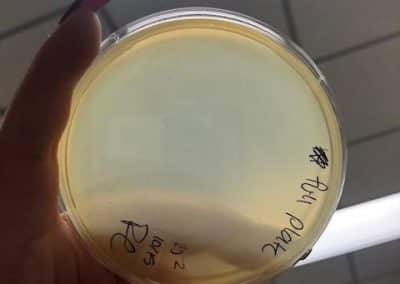
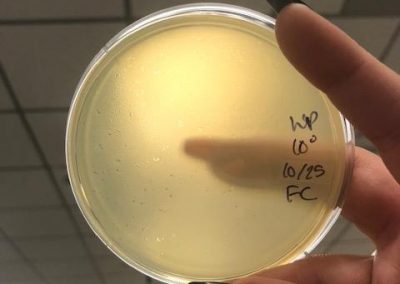
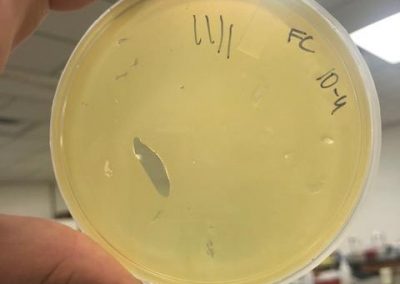
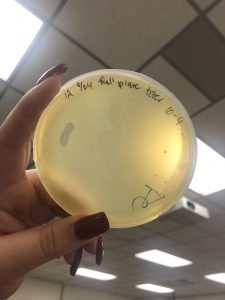
Photos taken on September 17, 2018

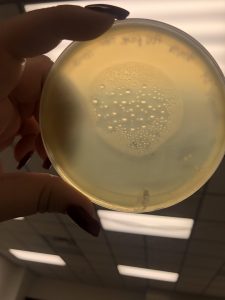
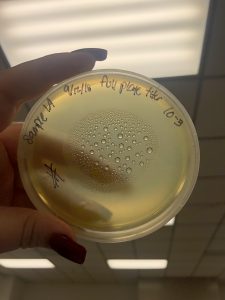
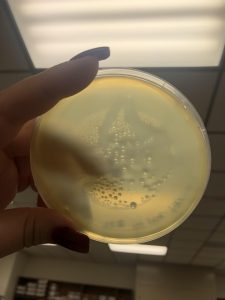
This was around when the phage buffer was contaminated. I threw out the samples used in the above plating from September 12, 2018.
September 24, 2018
- Serial Dilutions from Spot Titer
- Remade serial dilutions with my spot titer from September 10, 2018
- These serial dilutions were diluted down to 10^-4 and plated to make full plate titers
- Remade serial dilutions with my spot titer from September 10, 2018
- Full plate titers
- Prepared bacterial lawn plates for my dilutions to be used for full plate titers.
- Mixed 10 microliters of dilution with host bacteria and allowed the phage to attach for ten minutes.
- After attachment, each tube was mixed with 3 mL of top agar and plated.
- Incubated for a three days at 29 degrees Celsius
- The plates were contaminated 🙁
- Incubated for a three days at 29 degrees Celsius
Taken on October 1, 2018


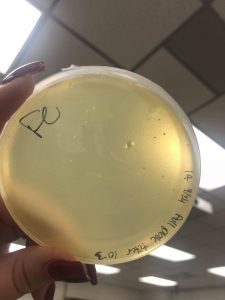
October 1, 2018
- Attempted another full plate titer
- mixed 10 microliters of phage dilutions with 250 microliter aliquots of host bacteria and allowed the phage to attach for ten minutes
- After attachment, 3 mL of top agar was mixed with the bacteria and plated.
- Incubated for four days (i forgot about it 🙁 )
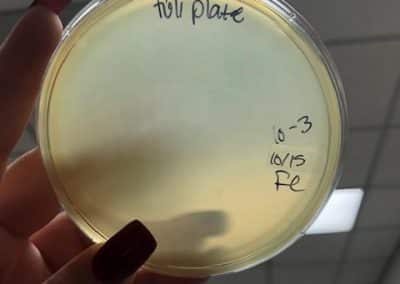
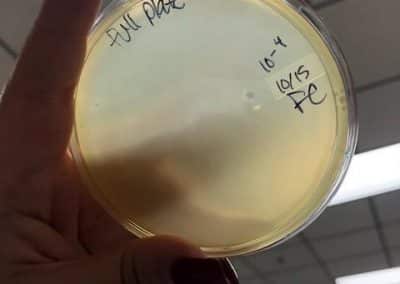
October 15, 2018
- Plated three full plate titers, 10^-2, 10^-3, and 10^-4. Plated those three because I believed they would yield the closest to a webbed plate without have an overabundance of phage.
- Plated using the standard full plate titer methods outlined in the SEA-PHAGES guide.
- allowed plates to come to room temperature. While coming to room temperature, I inoculated the host bacteria with 10 uL of my target dilutions.
- After around 20 minutes of attachment and warming up the plates, I plated the mixture of 250uL of host bacteria and 10uL of the target dilutions with 3mL of warm top agar.
- I allowed the top agar to set for around twenty minutes before flipping and incubating.
October 19, 2018
- Since Halloweenie was a slow growing phage ans clearly struggling, I let the plates grow for four days before giving up on those plates. The photos above were taken on October 19, 2018.
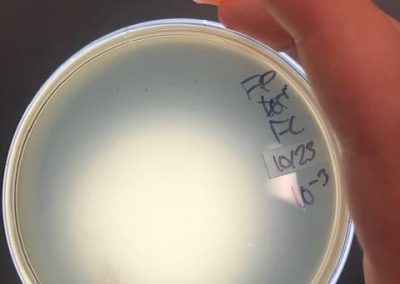
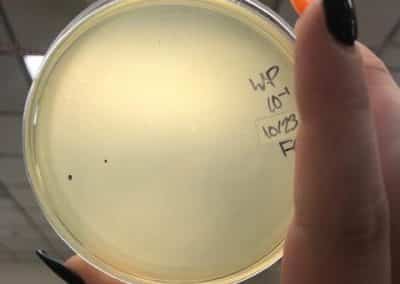
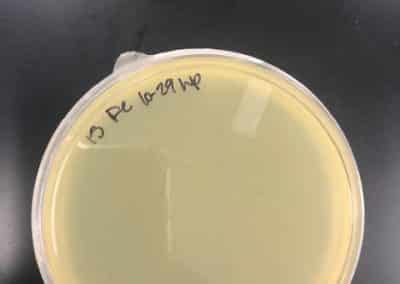
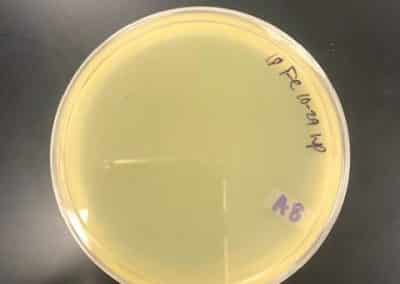
October 23, 2018
- After the weekend, I decided to try another set of full plate titers with a new set of serial dilutions.
- I went back to my plates from October 1, 2018, when I had a higher plaque concentration and a higher virulence than I was experiencing at the time, and picked a plaque.
- With this newly picked plaque, I redid my serial dilutions
- After redoing my serial dilutions, I tried another full plate titer/webbed plate without lysate with my fresh dilutions.
- Inoculated the host bacteria tubes with 10uL each of their predetermined dilutions. After inoculation, I allowed the phage to attach to its host for 15 minutes before plating with 3mL of top agar.
October 25, 2018
- Collected plate lysates
- Collected plate lysates by adding 6mL of phage buffer to my 10^-1 and my 10^0 plates. I averaged the number of plaques together when doing the math for my expected lysate.
- I attempted to make a webbed plate by plating 10uL of completely undiluted lysate. I left that to incubate for four days.
October 29, 2018
- Collected plate lysates
- collected the plate lysate from the above “webbed plate” by adding 8mL of phage buffer to the plate and letting it sit undisturbed on the counter in Genetics for three hours.
- After letting it sit for two hours, I gently swirled the plated for thirty seconds, to loosen phages from the agar and bacteria. Before propping the plate up on it’s lid and aspirating the lysate using a 5mL syringe.
- The syringe could only hold 5mL, so I aspirated 5mL and attached the syringe to the 0.22 um filter and dispensed the filtered lysate into a sterile 15mL conical tube.
- After all 5mL were dispensed, I removed the filter, careful not to tough the front of the filter, and placed it back in its container and used the same syringe to aspirate the remaining lysate and reattached the syringe to the previously used filter and dispensed the rest of the filtered lystate into the same 15mL conical tube as previously mentioned.
- This acted as my small volume lysate in future steps.
- Made webbed froms from a lysate of known titer to collect a high volume lysate
- Plated 15uL on 3 plates and 18uL on three plates of my small volume lysate that was collected on OCtober 29, 2018.
- plated 15uL and 18uL to increase my chacnes of getting a good webbed plate.
- Plated 15uL on 3 plates and 18uL on three plates of my small volume lysate that was collected on OCtober 29, 2018.
October 31st, 2018
- plated 25mL of my small voume lysate onto a plate to create a webbed plate that would be used to collect a high volume lysate on. Based on the concentration of my small volume lysate and the amount of plaques needed to created a webbed plate, I needed to plate 30uL of lysate, but plating more than 10% of the volume of my bacteria (250uL) could cause the cells to lyse.
November 1, 2018
- Picked a plaque from Rheaven’s original plaque assay from __________.
- Plaque was circled and marked with F
November 2, 2018
- Plated full plate titers using serial dilutions that Rheaven prepared on _____________ of phage D to make full plate titers to ensure that I am working with an isolated phage.
- these serial dilutions were taken from her full plate titers, meaning it still needed to go through one round of purification before it can be used for microscopy or archiving
- Also picked a plaque from the full plate titers of phage F from November 1, 2018
- Made more full plate titers from phage F to ensure the purity of plaques and that there is only one phage present.
November 4, 2018
- Collected a small volume lysate from the closest webbed plates of phage D.
- Collected a small volume lysate from the closest webbed plate of phage F.
- Also made a spot titer to determine the titer of the lysate collected from the F plates after they grew.
- Plated 20uL of my small volume lysate of phage F, and decided to stop working with phage D, as it grows more slowly and the concentration is lower.
November 5th, 2018
- Collected a high volume lysate that had a titer of __________________________.
- did electron microscopy
November 6th, 2018
- Zinc chloride DNA extraction
- Procedure:
- Gently mixed 5mL of the high volume lysate collected on November 4th, 2018 by inverting it three times.
- Added 20uL of nuclease mix
- Incubated the lysate/nuclease mixture for 10 minutes at 37 degrees Celsius
- Divided the mixture up among 5 1.5mL microcentrifuge tubes so they all had roughly 1mL of sample each.
- added 20uL of ZnCl2 to each tube and then mixed it by inverting the tubes 3 times and incubated them at 37 degrees Celsius for five minutes to precipitate the phage out.
- Centrifuge the tubes at max speed in the centrifuge near the autoclave in the room between genetics and the biochem lab for 1 minute.
- Remove the supernatants by aspirating. Kept the supernatant in a tube labels ZnCl2 supernatant
- resuspended the pellet in 500uL of TES buffer and incubated in 60 degres Celsuis for 15 minutes.
- Added 1uL of Proteinase K and gently mixed by inverting three times.
- Incubated the tubes at 37 degrees Celsius for 10 minutes
- Added 60uL of potassium actetate to each tube and mixed by inverting three times. Left on ice for 15 minutes.
- Centrifuged at max speed in the same centrifuge as earlier for 1 minute.
- Decanted the supernatant into new, labelled tubes.
- kept the tubes containing the capsid, just in case.
- Added 500uL of freshly prepared 80% isopropanol to each of the tubes and left them on ice overnight.
- Procedure:
November 7th, 2018
- Continued on the day 2 part of DNA extraction using the Zinc Chloride method
- Procedure
- centrifuged the tubes at top speed for 10 minutes
- Add 250uL of freshly prepared 70% ethanol into each tube and spun at top speed for one minute.
- Aspirated the ethanol out of the tube using a micropipette while visualizing the pellet the while time to ensure that I would not aspirate the DNA out and lose it.
- the supernatant was kept in a tub labelled as ethanol waste and was kept.
- allowed the DNA to dry for 12 hours at room temperature while sitting on a paper towel.
- Resuspended the DNA in 100uL of water and checked the concentration and quality of the DNA on the nandrop
- I didnt record the values on the nanodrop, but it was too salty and had RNA contamination.
- Procedure
- Purified my DNA sample using a ProMega Wizard PCR and gel extraction purification kit
- will insert protocol after i google it
- insert the photo of the nanodrop
- After talking with Dr. Edwards, we decided that my DNA extraction may have not been successful and I decided to try a few more methods of DNA extraction.