Discovery of JayKay
Collecting Soil Samples Date:9/03/17
Purpose: Collect soil samples from environments in which we may find a bacteriophage to infect Mycobacterium smegmatis. These samples will then undergo direct and enriched isolation to observe if the soil contained phage. Collect data on the soil collector, location description, general location, specific location, sample type, sample depth, and the ambient temperature.
Used protocol 5.1 to collect enviroment samples in various location. Scooped about 10-15mL of soil into three 50mL tubes. Recorded data onto a table for future reference.
Environmental Sample Data Sheet
32.3383° Lat
-96.9299° Long
32.3381° Lat
-96.9301° Long
32.2929° Lat
-96.9133° Long
Direct and Enriched Isolation started Date: 9/06/17
Began with protocol 5.2, the direct isolation of sample 3. The objective is to extract phages from the sample. Transferred 1/3 of the sample into a 15mL tube and then added enrichment buffer until the sample was submerged by 2-3 mL. Mixed thoroughly then incubated for 2 hours starting at 10:45 am. Removed then allowed particles to settle for 20 minutes. Attached a syringe to a .22µL fitler and removed the plunger. 2mL of the top liquid sample is added to the open syringe. Then the plunger is reinserted to filter out the sample into a microcentrifuge tube. Next I used protocol 5.3 for a plaque assay. I added 500µL of the direct sample into 250µL of M. smeg. Let sit for ten minutes in order for attachment. After ten minutes, I used a 5mL pipette to transport 3mL of top agar into the bacteria sample mixture. The quickly removed the full volume and transferred it onto an agar plate. Shifted the plate in multiple directions until the whole plate was coated. Placed in incubation.
Next, I proceeded to use sample 1 for the enriched isolation, protocol 5.5. The objective is to amplify phages present in the sample. Filled a 50mL tube with 15mL of sample. Added enrichment buffer up to the 35mL mark. Incubated from 10:40 am to 1:25 pm. After, sample was centrifuged four times until clear seperation. Clear liquid portion of sample is filtered through a .22µL vacuum filter to remove unwanted bacteria and soil particles. Added .5mL of M. smegmatis culture to filtrate. Placed back into incubation until next class.
Date: 9/08/17
Direct isolation assay and negative control assay are similar in appearance, illustrating no plaques are present in direct isolation. Enriched isolation removed from incubation.
Date: 9/11/17 Continuation of the Enriched Isolation
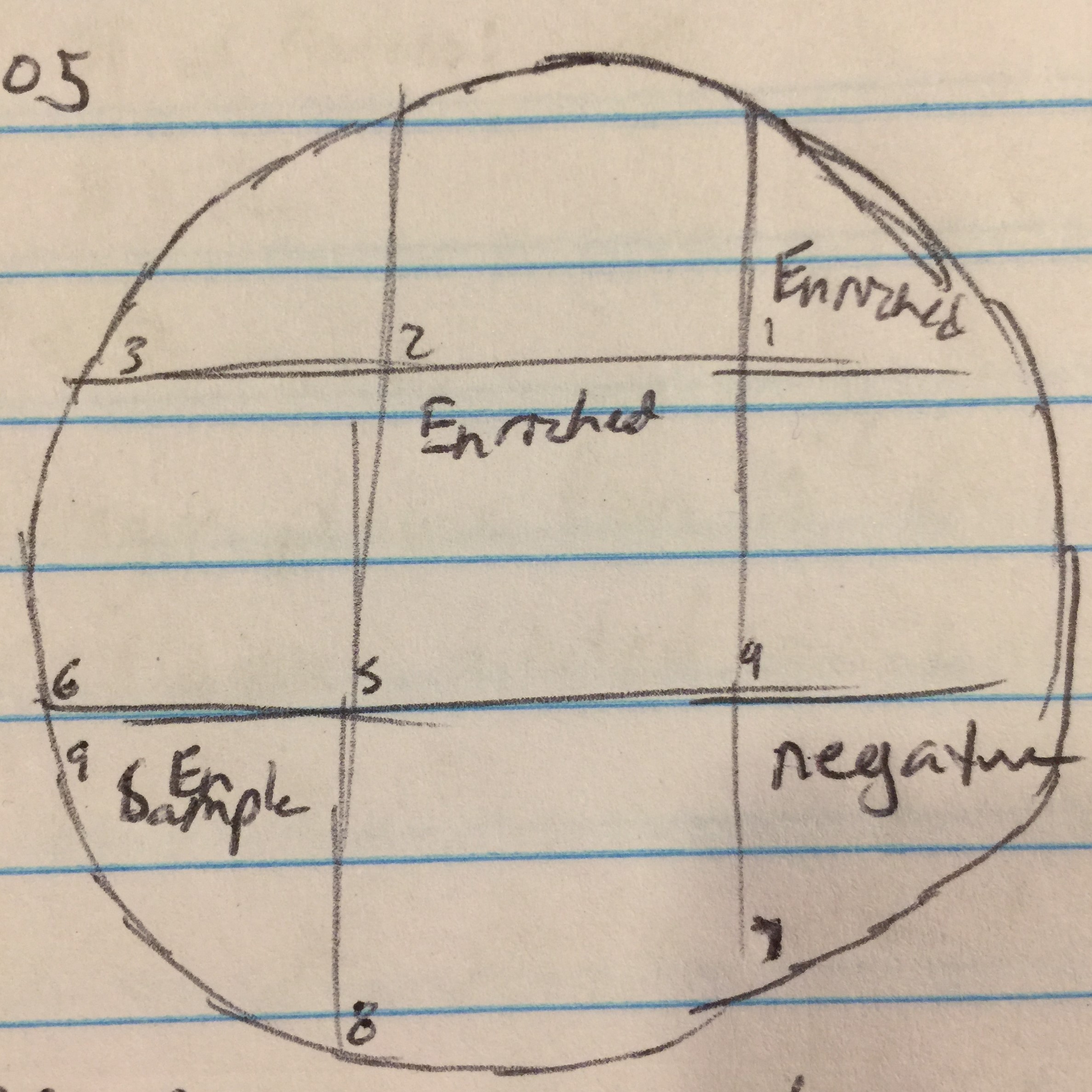
Day 2 of protocol 5.5. Transferred 1.4mL of enriched culture each to two microcentrifuge tube. Microcentrifuged for 1 minute. Took 1mL from each tube and filtered with .22µL filter into new tube. Proceeded to the spot test protocol. Used 250µl of host bacteria with 3ml of top agar to set the bacterial lawn. Allowed to sit undisturbed for 20 minutes until top agar solidified. Transferred 10µl of enriched sample to sections 1, 5, and 9. Section 7 was used as a negative control by dispensing 10µl of phage buffer. Incubated until 9/13/17.
Date: 9/13/17
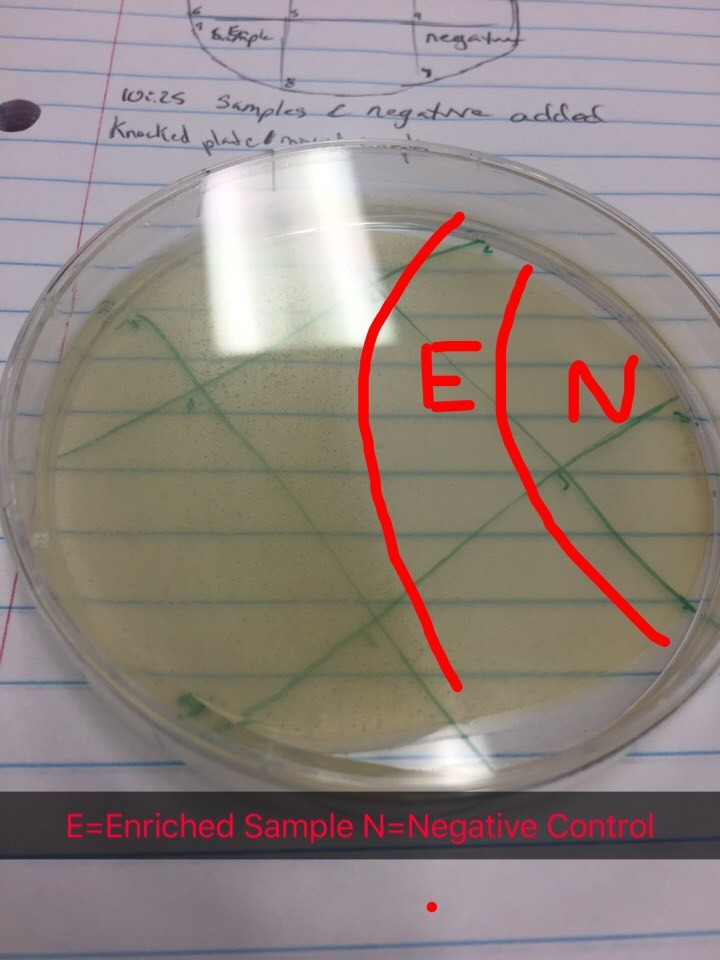
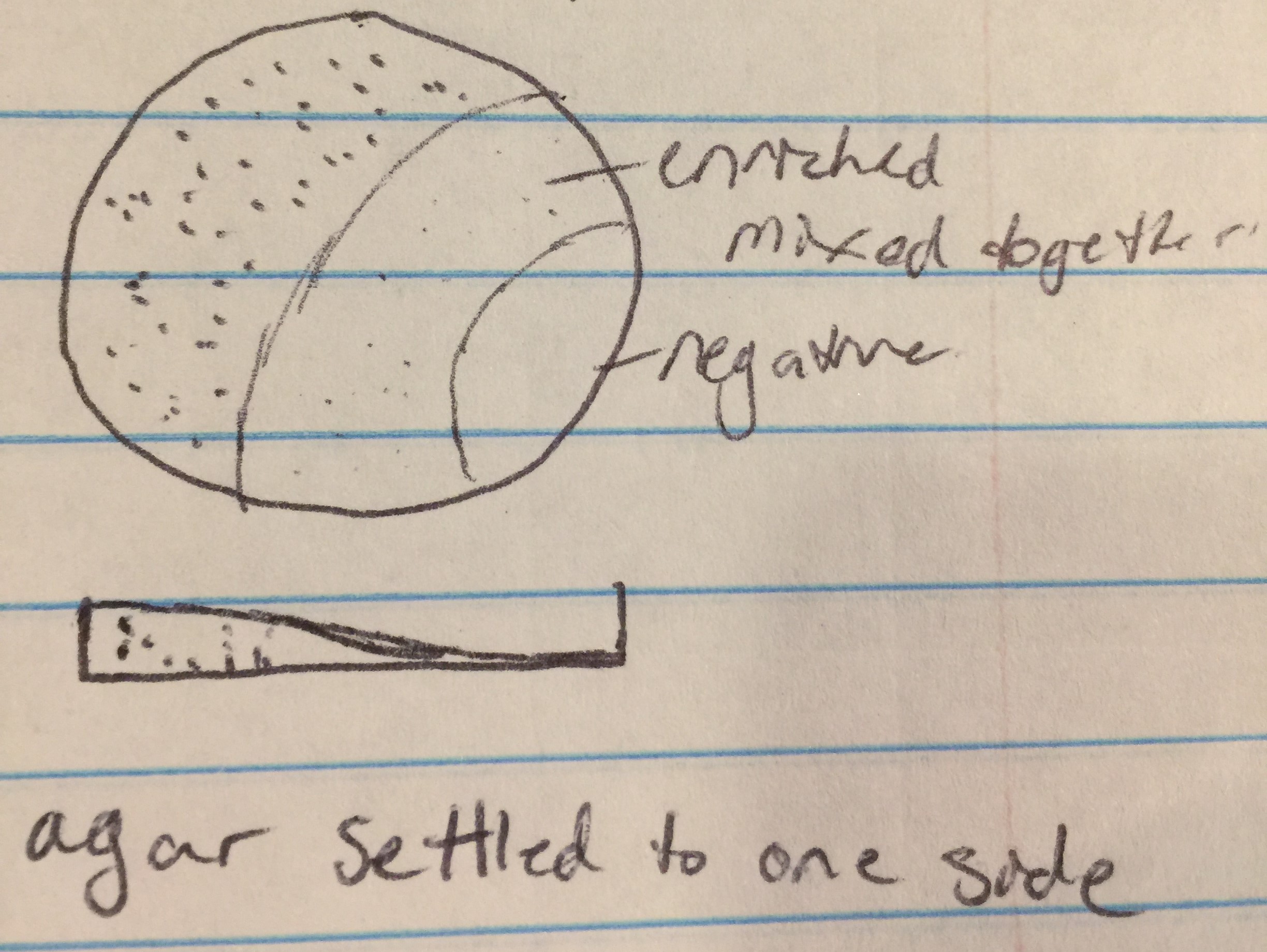
Results from spot test 1 show no phages. However, the agar was sloped, possibly causing problems for proper results.
Proceeded to do another spot test to ensure no phages are present in the enriched sample. Followed protocol 5.5 again. Top agar ended up with bubbles in all areas except 1, 4, 7, and 9. Transferred the enriched samples onto sections 1, 4, and 9. Section 7 was the negative control. Incubated for 48 hours.
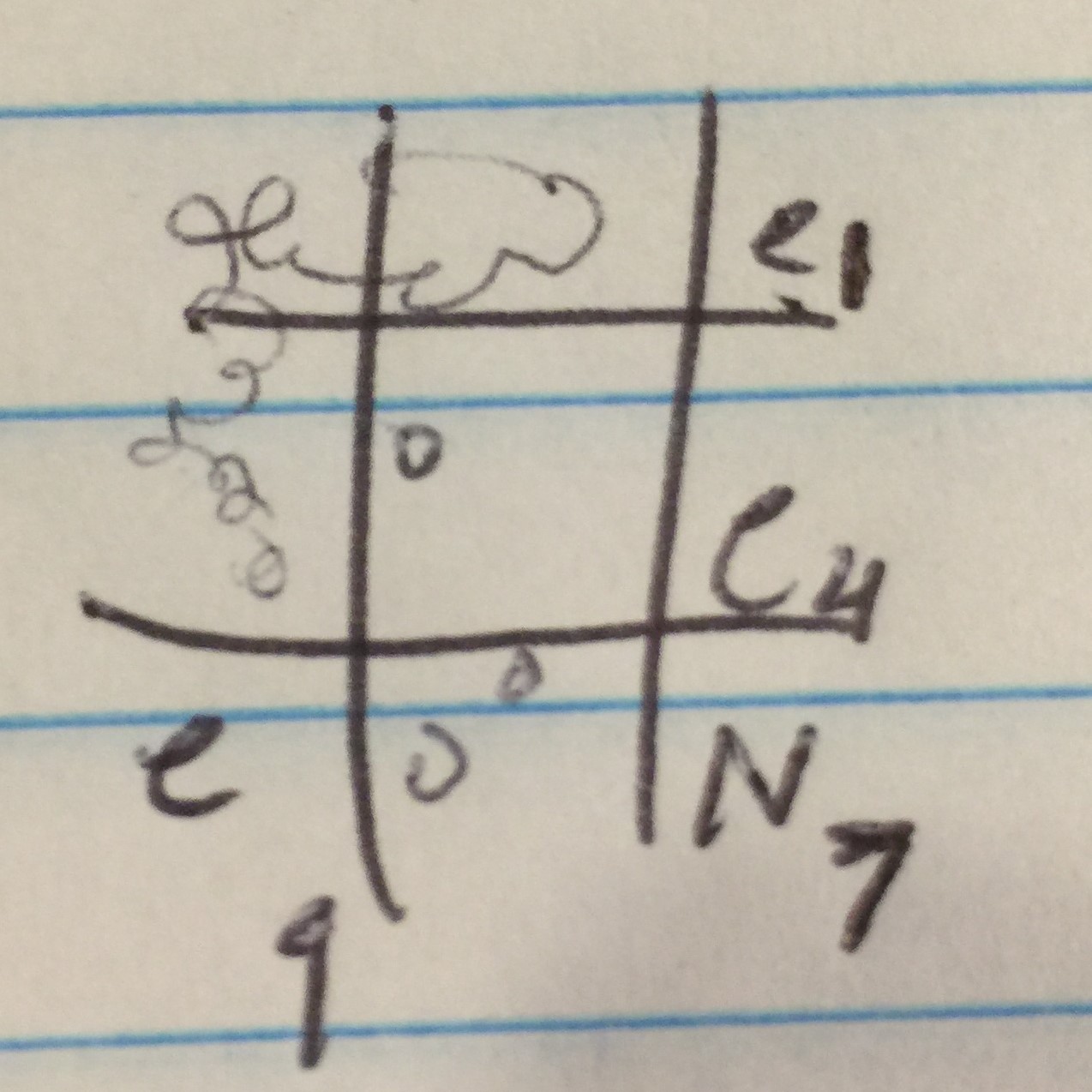
Date: 9/18/17
Spot test 2 had no phages. Adopted a direct isolation by picking a plaque from Bianca Willis’ plate. Location of adoption:
- Specific Location: 32.224998, -98.246089
Transferred 100µl of phage buffer into a microcentrifuge tube. Then “picked” a plaque by gently stabbing the top agar perpendicular to the surface with a p200 micropipetter. Placed the end of the tip into the microcentrifuge tube and proceeded to move around in the phage buffer to dislodge phages. Mixed by vortexing then started the serial dilutions. Serial dilutions are used to have samples of decreasing concentration of phage. Set up 6 microcentrifuge tubes, labeling from 10^-1 to 10^-6. Added 90µl of phage buffer to each tube. Added 10µl of the adopted direct isolation into the 10^-1 tube and vortexed. Then took 10µl of the 10^-1 sample and deposited it into the 10^-2 tube and vortexed. Repeated up until 10^-6. Plated each serial dilution using protocol 5.3 for plaque assays. Was taping plates together and rocked them causing the top agar to move. Placed in incubation until 9/20/17.
Date:9/20/17
Every assay was effected by the rocking of the top agar from the previous session. The 10^-1 assay showed plaques all around the outside of what seems to be agar that cooled or rippled unlike the rest.
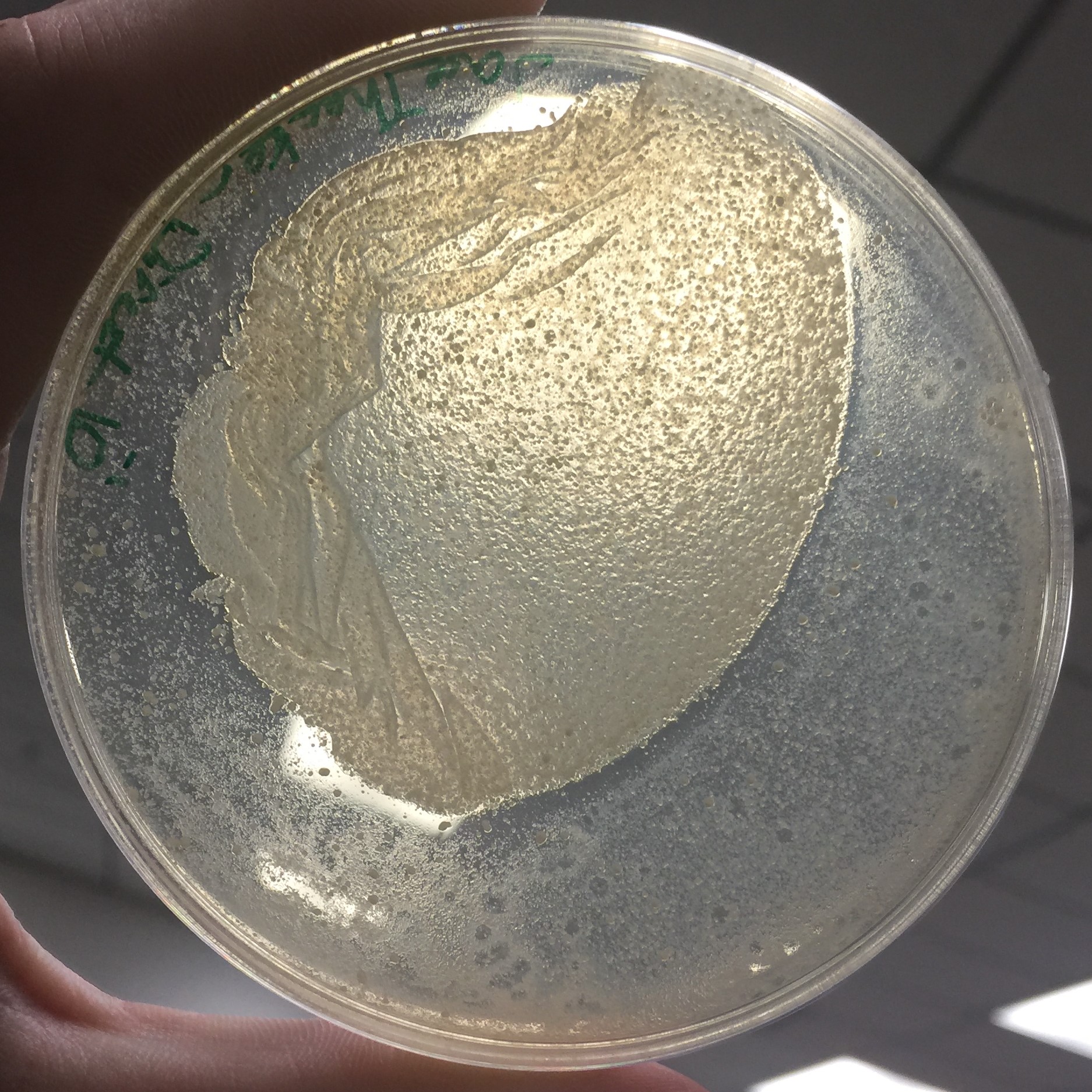
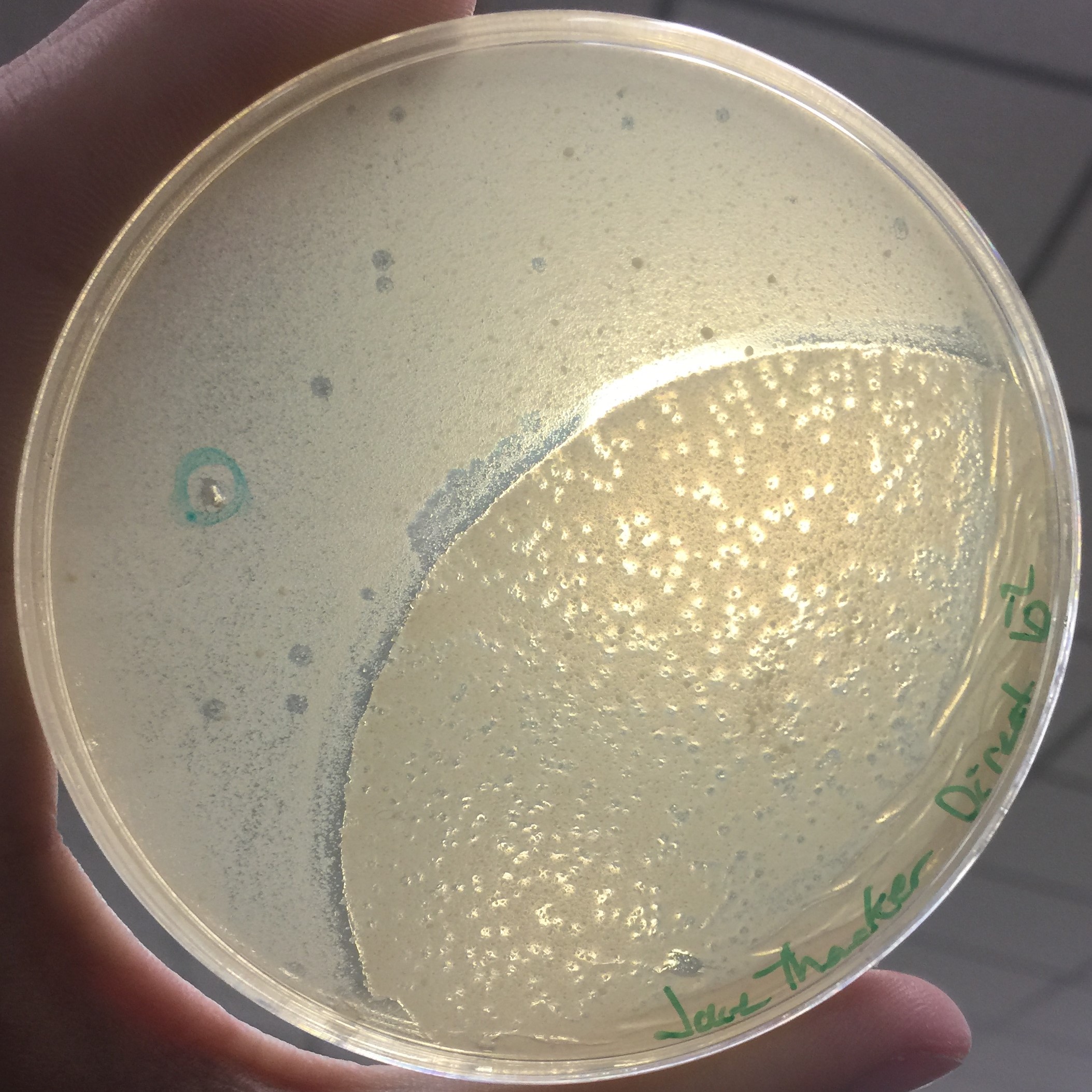
Results of Serial Dilution #2






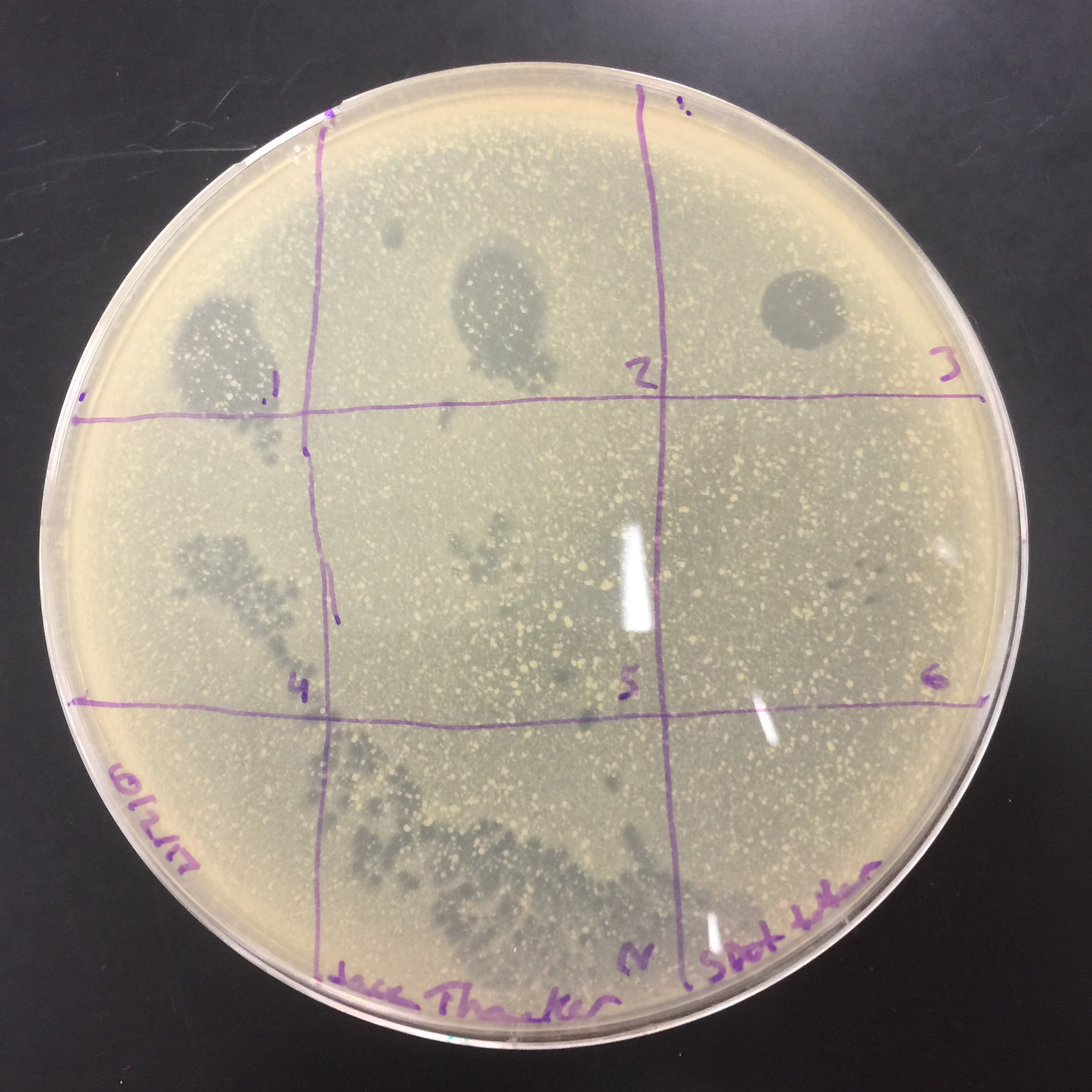
Results from the spot titer. Diltutions from 10^-1 to 10 ^-6 and a control in the bottom. From these results, I decided to take full plate titer for 10^-5 through 10^-7.
