Discovery of Allene
September 3, 2017
Title: Collecting Environmental Samples
Purpose: To collect soil and or water samples in order to find a bacteriophage that can infect the bacteria in those samples.
Procedure:
- A 50ml conical tube was labeled 1.
- Soil was collected in the tube, using the tube itself, and was filled approximately 3/4 full.
- Tube with soil sample was tightly capped and stored at room temperature (about 70 degrees Fahrenheit).
- A 50ml conical tube was labeled 2.
- Soil was collected in the tube, using the tube itself, and was filled approximately 3/4 full.
- Tube with soil sample was tightly capped and stored at room temperature (about 70 degrees Fahrenheit).
September 5, 2017
Procedure Cont.
- A 50ml conical tube was labeled 3.
- Soil was collected in the tube, using the tube itself, and was filled approximately 3/4 full.
- Tube with soil sample was tightly capped.
Results:
Conical Tube 1
- Location Description: A holding pen next to a hay bale in a horse/cow lot.
- General Location: A family farm/dairy on Co Rd 461.
- Specific Location: 32.209208, -98.300585
- Sample Depth: 1 inch
- Ambient Temperature: 84°F
- Physical Characteristics: Soil had some pieces of hay and grass, was dark brown in color, moist, and had an earthy smell.
Conical Tube 2
- Location Description: A horse lot next to water trough.
- General Location: A family farm/dairy on Co Rd 387.
- Specific Location: 32.208192, -98.288315
- Sample Depth:1 inch
- Ambient Temperature: 84°F
- Physical Characteristics:Soil had some pieces of grass, was light in color, sandy in texture, moist, and had an earthy smell.
Conical Tube 3
- Location Description: A goat/horse lot next to the water tank.
- General Location: A family home on the edge of Stephenville where suburbs turn into country.
- Specific Location: 32.224998, -98.246089
- Sample Depth: 1 inch
- Ambient Temperature: 62°F
- Physical Characteristics: Soil had some pieces of grass, was dark in color, contained some pieces of animal feces, and had an earthy scent.
Future Plans: To perform Direct Isolation Protocol and Enriched Isolation Protocol on the collected samples in order to find and collect bacteriophages.
September 6, 2017
Title: Enriched Isolation
Purpose: To amplify the bacteriophages present in soil sample previously collected.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A 50ml conical tube was filled to the 15ml mark with soil from conical tube 3.
- Enrichment broth was added to the 35ml mark, tube was capped tightly, and thoroughly vortexed/mixed.
- Tube was incubated at 37°C and shaken for 2hrs and 20 minutes at approximately 250rpm.
- Under a hood, supernatant was vacuum filtered and collected using a large .22um filter attached to a 50ml conical tube. 25ml of filtered supernatant was collected and tube was labeled.
- A laboratory burner was lit for aseptic work and bench wiped as before.
- 500ml of host bacteria was dispensed in the tube containing filtrate and incubated at 37°C and shaken at approximately 220 rpm for 2 days .
- After 2 days of incubation, tube was stored in refrigerator at 4°C.
Title: Direct Isolation
Purpose: To extract bacteriophages from a previously collected soil sample.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A 15ml conical tube was filled half way with soil from conical tube 3.
- Enrichment broth was added until soil sample was submerged under 4ml of liquid.
- The tube was capped and mixed thoroughly by shaking.
- The tube was incubated at 37 degrees Celsius and shaken for 2 hours and 20 minutes at approximately 250rpm.
- Sample was taken out and sat undisturbed until the matter settled at the bottom, about 10 minutes.
- A laboratory burner was lit for aseptic work and bench wiped as before.
- A syringe was taken apart, barrel and plunger separated.
- The barrel was attached to the .22um syringe filter and using a pipette filled with 3ml of supernatant, the top of the flooded sample.
- The plunger was attached back to the barrel, supernatant was filtered, and 1ml of filtrate was collected in a 1.5ml microcentrifuge tube.
- The 1.5ml microcentrifuge tube was labeled and then set aside.
Results:
1ml of filtered sample was obtained.
Future Plans: To perform Plaque Assay Protocol on the filtered sample obtained through Direct Isolation Protocol with sample 3.
Title: Plaque Assay
Purpose: To detect the presence of bacteriophages from the sample obtained through Direct Isolation Protocol done previously on a bacterial lawn.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit for aseptic work. All work forward is done under the “dome of heat”.
- A test tube rack and culture tube containing 250ul of host bacteria (Mycobacterium smegmatis) was obtained.
- With a pipette, 500ul of Direct Isolation sample was dispensed into the culture tube and mixed gently by swirling.
- Inoculated culture tube sat undisturbed for 10 minutes.
- Two agar plates and bottle of top agar was obtained and labeled (Direct Isolation Plaque Assay and the other Negative Control).
- With a sterile 5ml pipette, 3ml of top agar was dispensed in the culture tube containing host bacteria and direct isolation sample and immediately aspirated and mixture was transferred to the appropriately labeled agar plate.
- With a sterile 5ml pipette, 3ml of top agar was dispensed in the culture tube containing only host bacteria and immediately aspirated and mixture was transferred to the appropriately labeled agar plate.
- Agar plates was gently swirled and plates was mostly evenly coated.
- Small edge on left side of the Direct Isolation Plaque Assay plate lacked top agar.
- Negative Control plate had a bubble form in the top agar.
- Agar plates sat undisturbed for 15 minutes and top agar solidified.
- Plates was incubated for 2 days at 37°C.
- After 2 days of incubation, plates was parafilmed and stored in the refrigerator.
September 8, 2017
Results (Enriched Isolation Day 1): The sample was slightly cloudy but mostly clear, light yellow in color, and had bacteria clumped at the bottom of the 50ml conical tube.
Future Plans: To perform Day 2 of Enrichment Protocol.
Results (Plaque Assay):
- Plate had a small amount of large sized plaques with clearly formed edges.
- Some large plaque measurements: .25cm, .3cm, .4cm,
- Plate had a larger amount of small sized plaques, some with clearly formed edges and others with turbid edges.
- Some small plaque measurements: .05cm, .1cm
- Plaques are uniformly circular in shape.
Future Plans: To perform Picking a Plaque Protocol and Spot Test to confirm bacteriophages were present in the sample obtained from Direct Isolation.
September 11, 2017
Title: Enriched Isolation Cont.
Purpose: To extract bacteriophages that were amplified using culture bacteria in Day 1 of Enriched Isolation Protocol.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Two 1.5ml microcentrifuge tubes were obtained and labeled.
- 1.4ml of enriched culture, previously prepared, was dispensed in each microcentrifuge tube.
- Tubes were microcntrifuged at high speed for 1 minute to pellet the bacteria at the bottom.
- A syringe was taken apart, barrel and plunger separated.
- The barrel was attached to the .22um syringe filter and using a pipette filled with 2ml of supernatant, 1ml from each microcentrifuge tube.
- he plunger was attached back to the barrel, supernatant was filtered, and 1ml of filtrate was collected in a 1.5ml microcentrifuge tube and .4ml was collected in another 1.5 microcentrifuge tube. Tubes were labeled.
Results:
1.4ml of filtered supernatant was collected.
Future Plans: To perform Spot Test Protocol on the filtered supernatant collected.
Title: Picking a Plaque
Purpose: To obtain phage particles from the Plaque Assay for Direct Isolation and create a liquid sample to perform the Spot Test.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- With a marker, a plaque was marked for picking.
- A 1.5ml microcentrifuge was obtained and labeled.
- 100ul of phage buffer was dispensed into the tube.
- With a pipette and sterile tip, the top agar was lightly stabbed in the center of the marked plaque.
- The end of the tip was placed in the tube with phage buffer and the phage buffer was aspirated and dispensed gently, dislodging phage particles.
- Tube was vortexed and set aside.
Results:
100ul of phage in solution from a plaque was obtained.
Future Plans: To perform Spot Test Protocol on sample of phage in solution to confirm the presence of phages on the Direct Isolation Plaque Assay.
Title: Spot Test
Purpose:To confirm the presence of bacteriophages in the Plaque Assay (Direct Isolation) previously done and in sample obtained from Enrichment Isolation Protocol.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- An agar plate was obtained and labeled. The plate was sectioned into 9 sections.
- Section 1: Direct Isolation Phage Sample from Plaque Assay
- Section 3: Enriched Isolation Phage Sample (1)
- Section 5: Enriched Isolation Phage Sample (2)
- Section 7: Enriched Isolation Phage Sample (3)
- Section 9: Negative Control
- A test tube rack and culture tube containing host bacteria was obtained.
- With a sterile 5ml pipette, 3ml of top agar was dispensed into the culture tube containing host bacteria. Mixture was immediately drawn and dispensed on the agar plate.
- Plate was gently swirled to spread agar evenly. A small bubble was formed in section 4 and 7.
- Agar plate sat undisturbed until agar solidified, about 20 minutes.
- 10qul of each sample was aspirated and dispensed on its respective section on the agar plate.
- Plate sat for undisturbed for 40 minutes.
- Plate was incubated for 2 days at 37°C.
- After two days of incubation, plates were parafilmed and refrigerated.
September 15, 2017
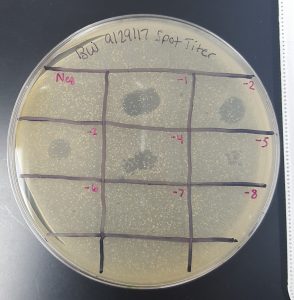
Results (Spot Test):
- There was cross contamination between section 1 and section 2 resulting in a large, smeared clearing on the top edge of the plate. It is uncertain which sample contained phages or not.
- Section 5 had a large clearing with distinct edges giving a positive result that phages are present in the sample obtained through Enriched Isolation Protocol.
- Section 7 had a medium size clearing with turbid edges giving a positive result that phages are present in the sample obtained through Enriched Isolation Protocol.
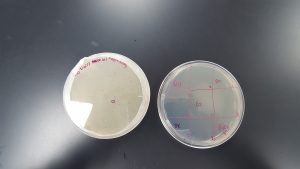
Future Plans: Using these results, it will be determined what samples will move forward and be subjected to Plaque Assay for Purification.
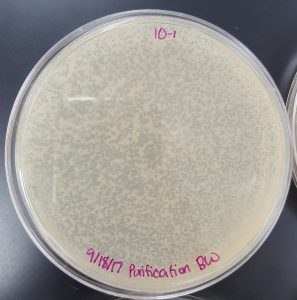
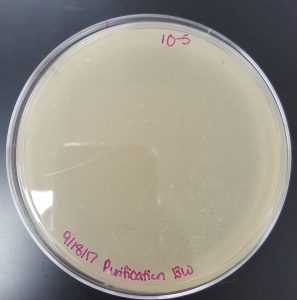
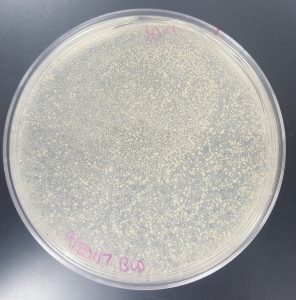
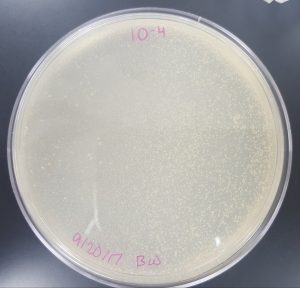
September 18, 2017
Title: Plaque Assay for Purification
Purpose: To produce isolated plaques from the Direct Isolation Plaque Assay.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- A plaque was picked and 100ul of phage in solution was obtained using the Picking a Plaque Protocol.
- The sample containing phage in solution was diluted using the Serial Dilution Protocol.
- Six agar plates were obtained and labeled 10^-1, 10^-2,…10^-6.
- Dilutions were plated according to the Plaque Assay Protocol. 10ul of each diluted sample was dispensed in its corresponding culture tube containing 250ul of host bacteria.
- Agar plate 10^-2 was knocked into which created a wrinkle/ripple in the top agar.
- Plates sat undisturbed for approximately 5 hours in order to allow top agar to set.
- Plates were incubated for 2 days at 37°C.
- After two days of incubation, plates are paraflimed and refrigerated.
September 20, 2017
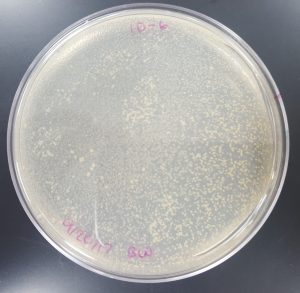
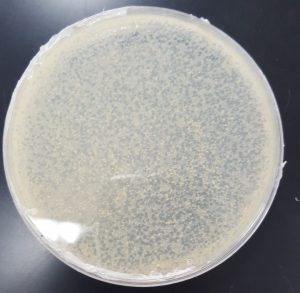
Results (Plaque Assay for Purification):
- 10^-1 Plate
- Plate contained a few thousand plaques which created a webbed pattern.
- Plaques are small and medium in size.
- Plaques are fairly circular and uniform in shape.
- Edges of the plaques are clearly formed.
- 10^-2 Plate
- A portion of the top agar was disturbed forming a wrinkle.
- There are a couple hundreds of plaques on the plate.
- Plaques are mostly medium in size with a few small in size.
- Plaques are fairly circular and uniform in shape.
- Edges of the plaques are clearly formed.
- 10^-3 Plate
- Plate has approximately 53 plaques.
- Plaques are mostly medium in size with a few small in size.
- The plaques are a little opaque and have turbid edges.
- Plaques appear fairly circular and uniform in shape.
- 10^-4 Plate
- Plate has 10 plaques.
- Plaques are small in size, opaque, and have turbid edges.
- Plaques appear fairly circular and uniform in shape.
- 10^-5 Plate
- Plate has 1 plaque.
- Plaque is small in size and has clearly formed edges.
- Plaque is circular.
- 10^-6 Plate
- Contains no plaques.
Future Plans: To perform Plaque Assay for Purification Protocol on one of the plates containing few phages.
Title: Plaque Assay for Purification (Round 2)
Purpose: To further isolate and genetically purify a plaque obtained in plate 10-4.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- A plaque was picked from the 10^-4 plate and 100ul of phage in solution was obtained using the Picking a Plaque Protocol.
- The sample containing phage in solution was diluted using the Serial Dilution Protocol.
- Six agar plates were obtained and labeled 10^-1, 10^-2,…10^-6.
- Dilutions were plated according to the Plaque Assay Protocol. 10ul of each diluted sample was dispensed in its corresponding culture tube containing 250ul of host bacteria.
- Plates sat undisturbed for approximately 7 hours in order to allow top agar to set.
- Plates were incubated for 2 days at 37°C.
September 25, 2017
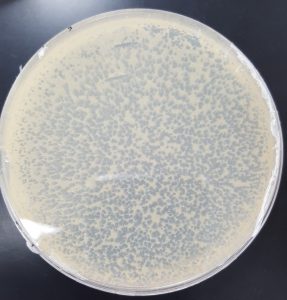
Results (Plaque Assay for Purification Round 2):
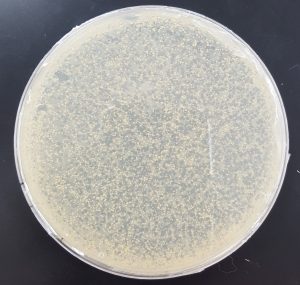
- 10^-1
- Plate has approximately 1000 plaques.
- Edges of the plaques are turbid, making the plate look almost clear.
- The plate is spotted white/yellow.

- 10^-2
- Plate has approximately 120 plaques.
- Plaques range from large to small in size.
- Edges of plaques are clearly formed.
- 10^-3
- Plate has 6 plaques.
- Plaques are medium and small in size.
- Edges of plaques are clearly formed.
- 10^-4
- Plate has no plaques.
- 10^-5
- Plate has no plaques.
- 10^-6
- Plate has no plaques.
Future Plans: To perform Collecting Plate Lysates Protocol on plate 10^-1.
Title: Collecting Plate Lysates
Purpose: To generate and collect a liquid sample with high concentration of phage.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- The 10^-1 plate was flooded with 8ml of phage buffer, swirled, and sat undisturbed for 4 hours at room temperature.
- After the incubation time was complete, the lid was removed and the edge of the plate was placed on the lid allowing the lysate to pool to one side.
- A .22um filter and 5ml syringe were opened and set aside.
- A 15ml concial tube was obtained and labeled.
- A 5ml syring was used to aspirate the lysate from the plate and was then attached to the .22um filter.
- The lysate was filtered and collected in a 15ml conical tube. Remaining lysate was filtered and collected as before into the same 15ml conical tube.
- The final volume of lysate collected was 5.50ml.
- Liquid phage sample was stored in the fridge at 4°C.
Results: 5.50ml of highly concentrated liquid phage sample was collected.
Future Plans: To perform Spot Titer Protocol on the phage sample collected.
September 29, 2017
Title: Spot Titer
Purpose: To determine the concentration of phage particles in the highly concentrated phage sample previously collected from a flooded 10^-1 plate after round 2 of purification.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- An agar plate was obtained and sectioned into 9 sections with a marker as well as labeled accordingly.
- Negative Control, 10^-1, 10^-2,…,10^-8. \
- A bacterial lawn was made using 250ul of host bacteria and 3ml of molten agar.
- Plate sat undisturbed until top agar was solidifed.
- The phage sample was diluted according to the Serial Dilutions Protocol (10^-1 through 10^-8).
- 3ul of control and serial dilutions were spotted onto the previously prepared bacterial lawn in the appropriate sections.
- Plate sat undisturbed until spots absorbed into the agar plate, about 2 hours.
- Plates were incubated right side up at 37°C for 2 days.
October 2, 2017
Results and Calculations (Spot Titer):
- The 10^-5 section showed 12 plaques small in size.
Titer(pfu/ml)= (#pfu/volume used in ul)x(10^3ul/ml)x(dilution factor)
Titer(pfu/ml)=(12pfu/3ul)x(10^3ul/ml)x(10^5)=4×10^8pfu/ml
Future Plans: To perform Making Webbed Plates from a Lysate of Known Titer Protocol on the highly concentrated phage sample.
Title: Making Webbed Plates from a Lysate of Known Titer
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- A: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Volume needed= (1.0×10^3pfu)/(4×10^8pfu/ml)=2.5×10^-6ul Therefore… 2.5ul of lysate from the 10^-6 dilution sample was added to 250ul of host bacteria.
- B:
- Volume needed= (2.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=5.0×10^-3ul Therefore… 5.0ul of lysate from the 10^-6 dilution sample was added to 250ul of host bacteria.
- A: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Sample was diluted according to the Serial Dilutions Protocol.
- Two culture tubes containing host bacteria were obtained and labeled A and B.
- Labeled culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Two agar plates were obtained and labeled A and B.
- Inoculated samples were plated to their respective agar plate according to the Plaque Assay Protocol.
- Plates were incubated without inversion at 37°C.
October 4, 2017
Results (Making Webbed Plates from a Lysate of Known Titer): Forgot to multiply by 1000ul/ml to properly convert volumes therefore, some small plaques were observed on the agar plates rather than webbed plates.
Future Plans: To redo the experiment with the proper calculations.
Title: Making Webbed Plates from a Lysate of Known Titer REDO
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- A: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Volume needed= (1.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=2.5×10^-3ul Therefore… 2.5ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- B:
- Volume needed= (2.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=5.0×10^-3ul Therefore… 5.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- A: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Sample was diluted according to the Serial Dilutions Protocol.
- Two culture tubes containing host bacteria were obtained and labeled A and B.
- Labeled culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Two agar plates were obtained and labeled A and B.
- Inoculated samples were plated to their respective agar plate according to the Plaque Assay Protocol.
- Plates were incubated without inversion at 37°C.
October 9, 2017
Results(Making Webbed Plates REDO 1): Plates did not have plaques.
Future Plans: To increase the hypothesized amount of plaques needed.
Title: Making Webbed Plates from a Lysate of Known Titer REDO 2
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- C: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Volume needed= (4.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=10.0×10^-3ul Therefore… 10.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- D:
- Volume needed= (6.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=15.0×10^-3ul Therefore… 15.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- E:
- Volume needed= (8.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=20.0×10^-3ul Therefore… 20.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- C: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Sample were diluted according to the Serial Dilutions Protocol.
- Three culture tubes containing host bacteria were obtained and labeled C, D, and E.
- Labeled culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Three agar plates were obtained and labeled C, D, and E.
- Inoculated samples were plated to their respective agar plate according to the Plaque Assay Protocol.
- Plates were incubated without inversion at 37°C.
October 11, 2017
Results (Making Webbed Plates REDO 2):
Plates dried up and showed signs of contamination (white fuzzy spots and splotches).


Future Plans: To redo making webbed plates and to cover the plates in parafilm in order for them not to dry out. Also, place them in the older incubator.
Title: Making Webbed Plates from a Lysate of Known Titer REDO 3
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- F: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Volume needed= (8.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=20.0×10^-3ul Therefore… 20.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- G:
- Volume needed= (10.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=25.0×10^-3ul Therefore… 25.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- H:
- Volume needed= (12.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=30.0×10^-3ul Therefore… 30.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- F: Volume needed=(number of plaques needed)/(titer of lysate)x(1000ul.ml)
- Sample was diluted according to the Serial Dilutions Protocol.
- Three culture tubes containing host bacteria were obtained and labeled F, G, and H.
- Labeled culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Three agar plates were obtained and labeled F, G, and H.
- Inoculated samples were plated to their respective agar plate according to the Plaque Assay Protocol.
- Plates were parafilmed closed to prevent drying out, placed in older incubated. and incubated without inversion at 37°C.
October 16, 2017
Results (Making Webbed Plates REDO 3):
- F
- G
- H
Future Plans: To collect lysate from all three plates.
Title: Collecting Plate Lysate
Purpose: To generate and collect a liquid sample with high concentration of phage.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Plates F, G, and H were flooded with 8ml of phage buffer, swirled, and sat undisturbed for 4 hours at room temperature.
- After the incubation time was complete, the lids were removed and the edge of the plates were placed on the lids allowing the lysate to pool to one side.
- Three .22um filters and 5ml syringes were opened and set aside.
- Three 15ml concial tube were obtained and labeled F, G, and H.
- A 5ml syring is used to aspirate the lysate from the plate and is then attached to the .22um filter.
- The lysate was filtered and collected in a 15ml conical tube. Remaining lysate was filtered and collected as before into the same 15ml conical tube.
- The previous two steps are repeated for G and H.
- Liquid phage samples were stored in the fridge at 4°C.
Results:
- F: 4ml of lysate was collected.
- G: 6.5ml of lysate was collected. The plate had a lot of moisture prior to flooding causing an increase of lysate collected compared to F and H.
- H: 4ml of lysate was collected.
October 31, 2017
Title: Phage DNA Extraction
Purpose: To obtain and isolate DNA from bacteriophage for Restriction Enzyme Digests and Sequencing.
Protocol:
- DNA was extracted from sample H according to the Phage DNA Extraction Protocol.
Results: 100ul of phage DNA sample were obtained.
Future Plans: To quantify the phage DNA obtained.
Title: Qubit Quantification
Purpose: To quantify the phage DNA sample previously obtained through Phage DNA Extraction Protocol.
Protocol:
- Three 0.5ml tubes for standards and samples were obtained, labeled, and placed on a rack.
- 597ul of Qubit dsDNA BR Buffer and 3ul of Qubit dsDNA BR Reagent were placed in 1.5ml microtube and lightly vortexed.
- 190ul of the master mix and 10ul of the appropriate Qubit standard were added to each tube used for standards.
- 199ul of the master mix and 1ul of the phage DNA sample were added to a previously labeled tube.
- All three tubes were lightly vortexed and incubated at room temperature for 2 minutes.
- Tubes were read using a Qubit 3.0 Fluorometer.
Results: 12.7 ng/ul was the concentration determined for the phage DNA sample.
Future Plans: To redo Phage DNA Extraction Protocol with some modifications.
November 2, 2017
Title: Phage DNA Extraction (REDO 1)
Purpose: To obtain and isolate DNA from bacteriophage for Restriction Enzyme Digests and Sequencing with some modifications to the protocol.
Protocol:
- DNA was extracted from sample F according to the Phage DNA Extraction Protocol with the following modifications.
- 10ul of nuclease instead of 5ul
- 15ul of EDTA
Results: 100ul of phage DNA sample were obtained.
Future Plans: To quantify the phage DNA obtained.
Title: Qubit Quantification
Purpose: To quantify the phage DNA sample previously obtained through Phage DNA Extraction Protocol.
Protocol:
- Three 0.5ml tubes for standards and samples were obtained, labeled, and placed on a rack.
- 597ul of Qubit dsDNA BR Buffer and 3ul of Qubit dsDNA BR Reagent were placed in 1.5ml microtube and lightly vortexed.
- 190ul of the master mix and 10ul of the appropriate Qubit standard were added to each tube used for standards.
- 198ul of the master mix and 2ul of the phage DNA sample were added to a previously labeled tube.
- All three tubes were lightly vortexed and incubated at room temperature for 2 minutes.
- Tubes were read using a Qubit 3.0 Fluorometer.
Results: 3.58 ng/ul was the concentration determined for the phage DNA sample.
Future Plans:To redo Phage DNA Extraction Protocol with some modifications.
November 6, 2017
Title: Phage DNA Extraction (REDO 2)
Purpose: To obtain and isolate DNA from bacteriophage for Restriction Enzyme Digests and Sequencing with some modifications to the protocol.
Protocol:
- DNA was extracted from sample F according to the Phage DNA Extraction Protocol with the following modifications.
- 10ul of nuclease instead of 5ul
- Lysate nuclease mixture was incubated for 15 minutes instead of 10.
Results: 100ul of phage DNA sample were obtained.
Future Plans: To quantify the phage DNA obtained.
Title: Qubit Quantification
Purpose: To quantify the phage DNA sample previously obtained through Phage DNA Extraction Protocol.
Protocol:
- Three 0.5ml tubes for standards and samples were obtained, labeled, and placed on a rack.
- 597ul of Qubit dsDNA BR Buffer and 3ul of Qubit dsDNA BR Reagent were placed in 1.5ml microtube and lightly vortexed.
- 190ul of the master mix and 10ul of the appropriate Qubit standard were added to each tube used for standards.
- 189ul of the master mix and 2ul of the phage DNA sample were added to a previously labeled tube.
- All three tubes were lightly vortexed and incubated at room temperature for 2 minutes.
- Tubes were read using a Qubit 3.0 Fluorometer.
Results: 5.79 ng/ul was the concentration determined for the phage DNA sample.
Future Plans: To use a different protocol recommended by the instructor.
November 8, 2017
Title: Making Webbed Plates
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- Volume needed= (16.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=40.0×10^-3ul Therefore… 40.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- Sample was diluted according to the Serial Dilutions Protocol.
- Three culture tubes containing host bacteria were obtained.
- Culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Three agar plates were obtained and labeled.
- Inoculated samples were plated to agar plates according to the Plaque Assay Protocol.
- Plates were parafilmed closed to prevent drying out, placed in older incubated. and incubated without inversion at 37°C.
November 10, 2017
Results (Making Webbed Plates):
Title: Collecting Phage Lysate
Purpose: To generate and collect a liquid sample with high concentration of phage.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- The three webbed plates were flooded with 8ml of phage buffer, swirled, and sat undisturbed for 4 hours at room temperature.
- After the incubation time was complete, the lids were removed and the edge of the plates are placed on the lids allowing the lysate to pool to one side.
- A .22um filters and 5ml syringe were opened and set aside.
- A 15ml conical tube was obtained and labeled.
- A 5ml syringe was used to aspirate the lysate from the plate and was then attached to the .22um filter.
- The lysate was filtered and collected in a 15ml conical tube. Remaining lysate was filtered and collected as before into the same 15ml conical tube.
- The previous two steps were repeated for the rest of the two plates.
- Liquid phage sample was stored in the fridge at 4°C.
Results: Fresh liquid phage sample was obtained.
November 15, 2017
Title: Making Webbed Plates
Purpose:To create a plate with a high density of plaques from a highly concentrated phage sample previously collected of known titer.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- Using a previous plaque assay, it was calculated/estimated how many plaques it takes to fill a plate.
- It was guessed it would take 1000 plaques or 2000 plaques to make a webbed plate.
- Calculations:
- Volume needed= (16.0×10^3pfu)/(4×10^8pfu/ml)x(1000ul/ml)=40.0×10^-3ul Therefore… 40.0ul of lysate from the 10^-3 dilution sample was added to 250ul of host bacteria.
- Sample was diluted according to the Serial Dilutions Protocol.
- Four culture tubes containing host bacteria were obtained.
- Culture tubes were inoculated with the appropriate volume from the appropriate dilution calculated above of liquid phage sample.
- Culture tubes were incubated for 10 minutes.
- Four agar plates were obtained and labeled.
- Inoculated samples were plated to agar plates according to the Plaque Assay Protocol.
- Plates were parafilmed closed to prevent drying out, placed in older incubated. and incubated without inversion at 37°C.
Title: Phage DNA Extraction Baylor Protocol
Purpose: To obtain and isolate DNA from bacteriophage for Restriction Enzyme Digests and Sequencing.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- 10ml of filter-sterilized phage lysate was transfered into a 50ml conical tube.
- 40ul of Nuclease Mix was added and the tube was mixed gently through repeated inversions.
- 4ml of PEG 8000/NaCl was added to the nuclease-treated lysate and tube was inverted repeatedly for gentle mixing.
- Solution was incubated for 30 minutes at 37 degrees Celsius.
November 16, 2017
- Tube was placed in a swinging bucket centrifuge and spun at 4100 rpm for 1 hour.
- Supernatant was decanted into the sink.
- 0.5ml of sterile water was added to tube and pellet was gently pipetted up and down to resuspend.
- 2ml of DNA Clean up Resin was added to the resuspended pellet and phage particles were uncoated by gently pipetting up and down. Tube was swirled to mix.
- Resuspended pellet with resin was distributed between two 1.5ml microtubes, 1.5ml per tube.
- Each tube was spun at 10000g for 6 minutes. Supernatant was removed.
- 1ml of freshly prepared 80% isopropanol was added to each tube. Pellet was washed by gently flicking the tube to mix.
- Each tube was spun at 10000g for 6 minutes. Supernatant was removed.
- Isopropanol wash was repeated.
- A 5ml syring was obtained,the plunger removed, and the barrel was attached to a column.
- Contents of both 1.5ml tubes were pipetted into the barrel-column and the plunger replaced pushing the flow through the column.
- Column was placed in a 1.5ml microtube and centrifuged at 10000g for 6 minutes.
- Column was transfered to a clean 1.5 microtube and 100ul of sterile water at 80 degrees Celsius was added. Column-microtube sat for 1 minute at room temperature.
- Column-microtube was centrifuged for 1 minute at 10000g and purified phage DNA was eluted into the microtube.
Results: 100ul of phage DNA sample were obtained.
Future Plans: To quantify the phage DNA obtained.
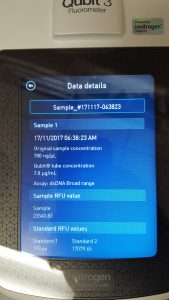
Title: Qubit Quantification
Purpose: To quantify the phage DNA sample previously obtained through Phage DNA Extraction Protocol.
Protocol:
- Three 0.5ml tubes for standards and samples were obtained, labeled, and placed on a rack.
- 597ul of Qubit dsDNA BR Buffer and 3ul of Qubit dsDNA BR Reagent were placed in 1.5ml microtube and lightly vortexed.
- 190ul of the master mix and 10ul of the appropriate Qubit standard were added to each tube used for standards.
- 198ul of the master mix and 2ul of the phage DNA sample were added to a previously labeled tube.
- All three tubes were lightly vortexed and incubated at room temperature for 2 minutes.
- Tubes were read using a Qubit 3.0 Fluorometer.
Results: 780 ng/ul was the concentration determined for the phage DNA sample.
Future Plans: To perform Setting Up Restriction Enzyme Digests Protocol on Phage DNA sample quantified.
November 17, 2017
Title: Collecting Phage Lysate
Purpose: To generate and collect a liquid sample with high concentration of phage.
Protocol:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- A laboratory burner was lit.
- The four webbed plates were flooded with 9ml of phage buffer, swirled, and sat undisturbed for 4 hours at room temperature.
- After the incubation time was complete, the lids were removed and the edge of the plates are placed on the lids allowing the lysate to pool to one side.
- Two .22um filters and two 5ml syringe were opened and set aside.
- A 15ml conical tube was obtained and labeled.
- A 5ml syringe was used to aspirate the lysate from the plate and was then attached to the .22um filter.
- The lysate was filtered and collected in a 15ml conical tube. Remaining lysate was filtered and collected as before into the same 15ml conical tube.
- The previous two steps were repeated for the rest of the three plates.
- Liquid phage sample was stored in the fridge at 4°C.
Results: Fresh liquid phage sample was obtained.
Title: Setting Up Restriction Enzyme Digests
Purpose: To cut phage genome into multiple fragments based on its DNA sequence.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- Tube containing DNA sample was lightly mixed by flicking it.
- Tube was incubated at 65 degrees Celsius for 10 minutes then placed on ice.
- The following enzymes were used: BamHI, ClaI, EcoRI, HaeIII, HindIII, SalI.
- Each enzyme reaction contained:
- 20.5ul of sterile water
- 2.5ul of 10X reaction buffer
- 1ul of restriction enzyme
- 1ul of phage genomic DNA
- For negative control:
- 24ul of sterile water
- 1ul of phage genomic DNA
- Contents of tubes were lightly vortexed and briefly centrifuged.
- Tubes were incubated at 37 degrees Celsius for 1 hour.
Title: Gel Electrophoresis of Restriction Enzyme Digests
Purpose: To separate DNA fragments via agarose gel electrophoresis.
Procedure:
- Agarose gel was prepared according to Casting Agarose Gels Protocol.
- 0.32g agarose powder
- 40ml 1X TBE buffer
- Gel was placed in the apparatus and apparatus was filled with 260ml of 1X TBE buffer.
- 2ul of 6x loading dye was added to 10ul of each restriction enzyme sample. 2ul of 6x loading dye was added to 5ul of 1kb ladder.
- Gel was loading in the following order: Ladder, Uncut DNA, BamHI, ClaI, EcoRI, HaeIII, SalI.
- Gel was run at 100V for 1 hour.
- Gel was stained using ethidium bromide/1X TBE buffer solution. Gel was then destained using 1X TBE buffer.
- Gel is visualized under UV light.
Results:
November 19, 2017
Title: Setting Up Restriction Enzyme Digests (REDO 1)
Purpose: To cut phage genome into multiple fragments based on its DNA sequence.
Procedure:
- Work bench was wiped down with 70% ethanol and other cleaning solution.
- Tube containing DNA sample was lightly mixed by flicking it.
- Tube was incubated at 65 degrees Celsius for 10 minutes then placed on ice.
- The following enzymes were used: BamHI, ClaI, EcoRI, HaeIII, HindIII, SalI.
- Each enzyme reaction contained:
- 20.5ul of sterile water
- 2.5ul of 10X reaction buffer
- 1ul of restriction enzyme
- 1ul of phage genomic DNA
- For negative control:
- 24ul of sterile water
- 1ul of phage genomic DNA
- Contents of tubes were lightly vortexed and briefly centrifuged.
- Tubes were incubated at 37 degrees Celsius for 1 hour.
Title: Gel Electrophoresis of Restriction Enzyme Digests (REDO 1)
Purpose: To separate DNA fragments via agarose gel electrophoresis.
Procedure:
- Agarose gel was prepared according to Casting Agarose Gels Protocol.
- 0.32g agarose powder
- 40ml 1X TBE buffer
- Gel was placed in the apparatus and apparatus was filled with 260ml of 1X TBE buffer.
- 2ul of 6x loading dye was added to 10ul of each restriction enzyme sample. 2ul of 6x loading dye was added to 5ul of 1kb ladder.
- Gel was loading in the following order: Ladder, Uncut DNA, BamHI, ClaI, EcoRI, HaeIII, SalI.
- Gel was run at 100V for 1 hour.
- Gel was stained using ethidium bromide/1X TBE buffer solution. Gel was then destained using 1X TBE buffer.
- Gel is visualized under UV light.
Restriction Enzyme Digests and Gel Electrophoresis Results:
November 20, 2017
Title: DNA Prep For Shipment
Purpose: To dilute the DNA sample from 780 ng/ul to 100 ng/ul for shipment to be sequenced.
Protocol:
- A labeled 1.5 microcentrifuge tube was obtained.
- 43.6ul of water was pipetted into the tube.
- 6.4ul sample was pipetted into the tube.
- Tube was lightly vortexed and briefly centrifuged.
Results: 50ul of DNA sample at 100 ng/ul.
November 27, 2017
Title: Archiving Your Phage Sample
Purpose: To prepare high-titer lysate for long term storage.
Protocol:
- High titer lysate was archived according to the Archiving Your Phage Sample Protocol except the following volumes were used: .
- 1.4ml high titer lysate and 100uL DMSO divided into 3 tubes with glass beads at 400uL in each tube.
Results: High titer lysate was archived.