Discovery of Sniffles
Environmental Samples
Sample 1 was a soil sample collected from Emporia County Compost Station, four inches beneath the surface. Collected on 08/30/2017 at 37.813199, -96.844323. According temperature at time of collection was 85 degrees F.
Sample 2 was a soil sample collected from a field surrounded by a subdivision, roughly three inches beneath the surface. Collected on 09/04/2017 at 31.0670 -97.6300. Recorded temperature at time of collection was 91 degrees F.
Sample 3 was a water sample collected from Belton Lake. Collected on 09/05/2017 at 31.1297, -97.5085. Recorded temperature at time of collection was 92 degrees F.
September 06, 2017
Direct Isolation preformed on Sample 1
- Soil sample filled to slightly above the 7 ml line.
- There was a larger chunk that sat on top of the sample that reached to the 8 ml line.
- Added 2 ml of enrichment broth
- Mixed thoroughly
- Incubated in a shaking incubator at 250 rpm from 11:26 AM until 1:11 PM
- Centrifuged sample for 10 minutes at 1000 RPM (1:33 PM to 1:44 PM)
No liquid was present after sample had been spun down. Repeated above steps with increased volume of enrichment broth.
Direct Isolation on Sample 1 (round 2)
- Soil sample filled to the 7 ml line.
- Chunks of soil were dumped onto table to avoid inaccurate measurements
- Added 3 ml of enrichment broth
- Mixed thoroughly
- Incubated in a shaking incubator at 250 rpm from 1:54 PM until 4:11 PM
- Centrifuged sample for 10 minutes at 1000 RPM (4:15 PM to 4:25 PM)
- Let sample sit for nine minutes to ensure particulate matter settled
- Filtered top liquid from sample then proceeded to plaque assay protocol
Plaque Assay
- Introduced 500µl of phage sample with 250 µl of host bacteria
- Vortexed sample and let sit from 5:07 PM until 5:28 PM
- Created negative control with 10 µl of phage buffer and host bacteria. Sat from 5:10 until 5:20
- Added 3 ml of agar to each sample and then plated
- Agar was not properly spread on negative control. Direct isolation sample was plated well.
- Plates were incubated for 24-48 hrs after sitting for roughly 20 mins
Incubation time for direct isolation: 5:25. Starting temperature at 37 degrees C
Incubation time for negative control: 5:42, Starting temperature at 37 degrees C
Enrichment isolation preformed on Sample 2
- Soil sample filled to the 15 ml mark in 50 ml test tube
- Enrichment broth filled to 35 milliliters (20 ml)
- Sample shook at 250 rpm from 10:55 AM until 12:57 PM
- Filtered sample and collected flow in new test tube
- When filtering sample, detached filter from the vacuum in order to let a classmate use while centrifuged sample
- Introduced 500 µl of host bacteria to sample
- Incubated at 220 rpm from 5:34 PM until
September 08, 2017
Removed Sample 1 form incubator at 11:07 AM @ 37 degrees C.
When viewing both of the plates, there is no dominant plaques visible. When held up to the light, there does not seem to be any sign of plaques showing up either. It does not seem like a plaque at this point. No pictures were taken. Further incubation took place to make sure sample was not possibly a slow grower. Placed back into incubator at 11:21 AM at 37 degrees C.
Was viewed 09/10/2017, no plaque present. Will be proceeding with the enriched isolation to see if phages are present.
September 11, 2017
Removed enriched isolation from incubator by professor then kept in the refrigerator. Filtered sample once more before plating.
Spot Test preformed on Sample 2
- Split plate into nine section
- Introduced agar with host bacteria then placed onto plate
- When placing agar onto plate, did not dispense quickly enough so did not seem to cover section 4 and 7 completely. These areas were avoided.
- Let plate sit from 9:51 AM until 10:13 AM
- Plate was moved by classmate during the 20 minute solidifying process. Negative sample crossed over into section 3, 6, and 5.
- Placed phage buffer into section 2
- Placed enriched sample (Sample 2) into sections 1 and 9
- Incubated at 10:39 AM at 37 degrees C
September 13, 2017
Cont’d spot test preformed on Sample 2
Spot Test was taken out of incubator at 9:10 AM.
In section 6 and 9, there was one sand sized yellow dot. This was contamination on plate. Agar plate was slightly dried out as well more towards section 1, 4, and 7. Agar also seemed to be shifted, with less agar present within sections 1, 4, and 7. Seemed to be an issue with rest of class as well. This led to the rest of the class redoing their spot samples to test accuracy. Due to the failure of the direct isolation of Sample 1 and enriched isolation of Sample 2, decided to restart from the point of direct isolation within lab manual, but with Sample 3.
Direct isolation preformed on Sample 3
- Immediately filtered sample through .22 µm filter due to it being a water sample
- Did two separate filtrations. Two different samples (Sample D1 and Sample D2) from Sample 3.
- Negative control vortexed with phage buffer, 9:27 AM. Plated with agar 9:48 AM
- First one that was plated so bubbles were formed, tried removing them. Seems as if I placed the end of the pipettor too far into agar. Two slight pokes in agar present. Water droplets sporadically all over this plate before incubation.
- Sample D1 not vortexed, left to settle on own. Added together with host at 9:57 AM. Plated at 10:23 AM.
- Best plated agar. No bubbles.
- Sample D2 vortexed. Added together with host bacteria at 10:05 AM. Plated at 10:25 AM.
- Was going to let sit longer, but got caught up in plating and just kept going. Two bubbles formed, but able to remove both of them.
- While agar was setting, plates were stacked together.
- Incubated Sample D2 and negative control at 11:03 AM
- Did not invert these plates until 11:28 AM.
- Let plate D1 sit until 11:27 AM then incubated (inverted)
September 15, 2017
Cont’d direct isolation preformed on Sample 3
10:37 AM
Sample D1 and D2 was checked. Nothing seemed to be growing within either sample. May be slow grower so letting stay over weekend. Checking Monday. Continued incubation at 37 degrees C. Decided to to use Brittany Stewart’s Direct Isolation with multiple phages present (vegetable garden soil sample from Bluffdale).
Picking a plaque
- Chose a plaque
- Brittany Stewart and Camille Trautman already chose plaques to further their research
- Chosen plaque was a significant amount of distance from other plaques
- Circled plaque with a green marker and labeled with initials “LH”
- Chosen plaque had a circular appearance, but upon closer inspection could see that the plaque was in the temperate stage due to it being turbid
- Plaque was measured at 0.18 cm
- Placed 100 µl of phage buffer into 1.5 ml tube and combined with picked plaque
Spot test preformed on adopted phage
- Obtained plate and split into nine sections
- Introduced host bacteria with agar and then immediately plated
- First agar plate made solidified quicker than expected and was inconsistent. Plated at 11:28 AM. Created second agar plate, solidified quickly but was evenly distributed besides section 1. Was not evenly distributed in the sense that I could not get the agar in that section within a decent amount of time. Let agar solidified for 21 mins (11:31 AM – 11:52 AM). Was going to use this plate for spot test for adopted phage, however; my thumb accidentally stabbed into the agar while I was trying to plate adopted phage. In order to avoid contamination and humiliation for life, obtained third plate, plated agar, and then plated adopted phage. Third agar plate sat from 11:58 AM – 12:21 PM.
- Plated 10µl of mixture created from Picking a Plaque protocol into each chosen section
- Section 2 was used for negative control
- Section 4, 6, 8 were used for direct isolation sample of adopted phage
- Slight splatter occurred in section 6
- Plated at 12:24 PM an incubated at 10:18 PM at 37 degrees C
When getting 250 µl of host bacteria, had to half sample containing 500 µl. This was because there was no sterile 250 µl samples available. Used micropipetter to split bacteria. Might be inaccurate because the pipetters measurements started at .5 ml. Eyeballed halfway from this mark to obtain 250 µl. Placed separated bacteria into another test tube. When comparing them to one another, seemed as if one had a slightly higher volume than the other. Greater volume was combined with phage. Phage may grow slower because of this.
September 18, 2017
Cont’d direct isolation preformed on Sample 3
The second round of direct isolation that was preformed showed no results. Rest of research will be preformed using adopted phage from Brittany Stewart’s direct isolation.
D1 sample D2 Sample
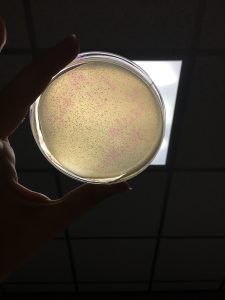
Brittnay’s Direct Isolation

Cont’d spot test preformed on adopted phage
- Clearings were observed in sections 4, 6, 7, and 9
- Clearings in section 4 and 6 were circular and did not smear into any other sections. Was controlled. Clearing in section 8 did cross over into section 9.
- Picture of results shown below
Plaque assay of dilution series
- Placed 100 µl of phage buffer into 1.5 ml tube and combined with picked plaque
- When trying to obtain the phage from the plate, was bumped into and caused the tip of the pipette to go into agar. This caused a tear in the agar of the chosen plaque. The tear was took over the circumference of the plaque. Chose another plaque on the plate for serial dilutions. Did preform a spot test for this new plaque. just followed through with next steps with new picked plaque.
- Set up six 1.5 ml test tubes with 90 µl of phage buffer in each
- Transferred 10 µl of plaque mixture into the first test tube
- Transferred 10 µl of first sample into the second test tube
- Followed out each dilution by taking 10 µl of the previous test tube until reached the last one
- Combined bacteria host with each dilution at 11:34 AM and let sit until 11:45 AM
- Started process of combining agar and dilution with bacteria at 11:56 AM
- Let plates sit before incubation until 4:01 PM
September 20, 2017
Checked plates and it seems that only 10^-1 had many plaques present. Pictures taken, shown below. All plaques have distinct circular shape. Are spread out somewhat evenly. No clustering was present. However, there was a couple plaques that had another plaques attached to it. Dilution 10^-3 seemed to not even have host bacteria present in it. Threw away all other plates that had nothing present on them.

Picked a plaque from 10^-1 dilution plate.
5:50 PM time serial dilutions were added to host bacteria.
Started plating at 6:02.
*Added all of serial dilution into the host bacteria. Instead of only adding 10 ul, added 100 ul.*
September 25, 2017
Plaque assay of dilution series cont’d
Pulled from incubator at 9:37 AM.
All dilutions seemed to have phage present besides 10^-6.
10^-1, 10^-2, 10^-3 seemed to have a tilt in the agar.
Pictures present below. Plates are labeled accordingly.
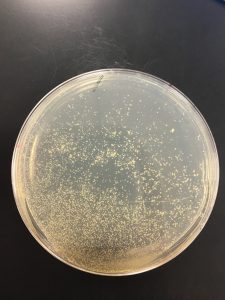
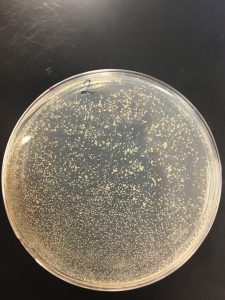
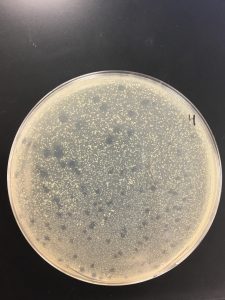
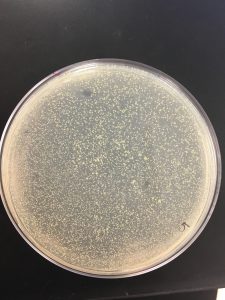
Collecting plate lysates
- Added 8 mL of phage buffer to 10^-4 at 11:10 AM.
- Let plate sit until 4:09 PM at room tempature
- Gently swirled solution before extracting liquid from plate
- Tilted plate slightly and used a syringe to extract lysate
- Filtered through a .22 um filter and placed into a 15 ml conical tube
Filtered lysates volume: 5.5 ml
Filtered at 4:23 PM.
Filtered lysates stored at 4°C : 4:29 PM.
September 28, 2017
Spot Titer
- Divided agar plate into nine sections
- Label one section as negative control and then the other sections in order from 10^-1 to 10^-8
- Combine 3 ml of top agar to 250 ul of host bacteria and immediately plate
- Let solidify completely
- Preform serial dilutions on lysate obtained from Collecting plate lysates protocol
- Use 10 ul of lysate and phage buffer for dituion
- Transfer 3 ul of each dilution into the correlating spot labeled on the plate
- Let sit until spots are completely absorbed into the plate
- Incubate for 24 – 48 hours
September 27, 2017
Spot Titer cont’d
Full plate titer
- Label eight different agar plates 10^-1 through 10^-8
- For each serial dilution created for Spot Titer protocol, combine 10 ul of it with 250 ul host bacteria
- Combine 3 ml of top agar with mixture previously created and plate it with the according labeled plate
- Let solidify then incubate for 24- 48 hours
September 2, 2017
Full plate titer cont’d
October 02, 2017
10/11/17
Found little results for full titer second round. Nothing on 10^-3. Two small plaques on 10^-4 and one on 10^-5. Pictures taken. Stored second round of dilutions in the initial direct and enriched solutions due to there being no more space in proper container.
Redid lysate serial dilutions. Did not preform spot titer. Took 10^-3 and 10^-4 to plate for full titer. \
Combined host and serial dilution at 11:58 AM. Added to agar then plated at: 12:23PM. Incubated at:
4:29
Side note: see if these two dilutions work out. And if not, go ahead and try to do the 10^-1 and 10^-2. Did have plaque on 10^-4 on spot titer but it may of also been 10^-1 crossing over into the section. So to be sure may need to try out 10^-1.
10/16/17
10^-3 dilution of lysate had 408 plaques
10^-4 diltuion of lysate had 39 plaques
Calculating titer in pfu/ml
(408 pfu / 20ul) (10^3) (10^5) = 2.04 x 10^9 pfu/ml
(39 pfu / 10 ul) (10^3) (10^5) = 3.9 x 10^8 pfu/ml
Decided that I had smaller plaques. Therefore, I based my volume needed off this assumption.
Calculating volume of lysate needed for webbed plate
(1.5 x 10^3 pfu) / (2.04 x 10^9 pfu/ml) = 7.35 x 10^-7 ml lysate
(7.35 x 10^-7 ml lysate) (1000 ul/ml) = 7.35 x 10^-4 ul
(1.5 x 10^3 pfu) / (3.9x 10^8 pfu/ml) = 3.85 x 10^-6 ml lysate
(3.85x 10^-6 ml lysate) (1000 ul/ml) = 3.85 x 10^-3 ul
Decided to try to bracket the number of phage particles that would be put onto plate. Therefore, also plated ten times more and ten times less than original calculations.
So plated 7.35 ul of the 10^-3 lysate dilution, 7.35 ul of the 10^-4 lysate dilution, and 7.35 ul of the 10^-5 dilution.
Also plated 3.85 ul of the 10^-2 lysate dilution, 3.85 ul of the 10^-3 lysate dilution, and 3.85 ul of the 10^-4 lysate dilution.
Combined the dilutions for 7.35 with 250 ul of host bacteria at 1:02 PM.
Combined the dilutions for 3.85 with 250 ul of host bacteria at 1:10 PM.
Let these sit until 4:12 PM then started plating each of them accordingly. Ended plating at 4:24 PM.
Incubated these plates until October 17, 2017 at 5:05 PM. Each plate had no results.
10/18/17
Looked over calculations again and realized that I’m slow af. When calculating the titer in pfu/ml I did not calculate my plaques by the correct dilution factor. Used the text books dilution factor instead. Also decided that my plaques were more medium sized than they were small. Furthermore, when calculating the volume of lysate needed to generate a webbed plate, increased the estimation of plaques needed significantly. Was not aware that this number had as much fluidity as it did. Redid calculations. Shown below.
10^-3 dilution of lysate had 408 plaques
10^-4 diltuion of lysate had 39 plaques
Calculating titer in pfu/ml
(408 pfu / 20 ul) (10^3) (10^4) = 3.9 x 10^7 pfu/ml
(39 pfu / 10 ul) (10^3) (10^3) = 20.4 x 10^6 pfu/ml
Calculating volume of lysate needed for webbed plate
(12000 pfu) / (3.9 x 10^7 pfu/ml) = 3.076 x 10^-4 ml lysate
(3.076 x 10^-4 ml lysate) (1000 ul/ml) = 3.076 x 10^-1 ul
(12000 pfu) / (20.4 x 10^6 pfu/ml) = 5.88 x 10^-4 ml lysate
(5.88 x 10^-4 ml lysate) (1000 ul/ml) = 5.88 x 10^-1 ul
Due to there needing to be more agar plates created, was only able to plate one dilution on this day. Combined 3.08 ul of dilution 10^-1 at 11:31 AM. Plated with 3 ml of agar at 11:41 AM. Let incubate until 10/20/17 at 9:45 AM.