Discovery of TidBit
August 30th, 2017
Instructed to collect three soil samples over the weekend. Followed Protocol 5.1: Collecting Environmental Samples.
September 6th, 2017
Performed Protocol 5.2: Direct Isolation and Plaque Assay Protocol 5.3 with Sample #1. Performed Protocol 5.5: Enriched Isolation with Sample #2.
Noted that I failed to properly maintain a portable flame lit around experiment area until Step C) 3). Dr. Edwards informed me of mistake along with suggestion I repeat preparation of Sample #1 Direct Isolation Attempt #1. Due to not enough useable suspension of material. Attempt #2 was completed according to Protocol 5.2: Direct Isolation and placed in shaking incubator at 16:27 C.T. While waiting to perform Plaque Assay of Sample #1, Sample #2 under went Protocol 5.5: Enriched Isolation. Enriched Isolation sample was placed in shaking incubator at 16:48 C.T.
Performed Protocol 5.3: Plaque Assay on Attempt #2 of Direction Isolation Protocol. Group was instructed to not have a positive control.
September 11th, 2017
Observed no plaques on Direct Isolation plates begin Spot test on Enriched Sample.
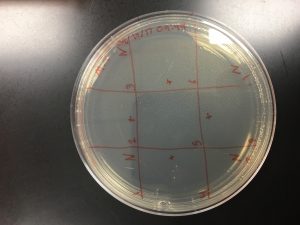
Performed Day 2 Protocol 5.5: Enriched Isolation with Sample #2. After enriched sample was prepared completed Protocol 5.6: Spot Test on E.I. Sample #2. Attempted a 3×3 pattern.
* N indicate negative control and + indicate placement of EI Sample #2
September 13th, 2017
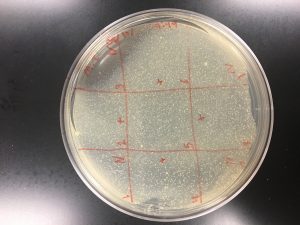
Observed no plaque formations on Spot Test Attempt #1. Instructed to attempt Spot Test again with own enrichment samples and those of other fellow classmates. Jonathan and Brandi both allowed me to use their enrichment samples for Spot Test Attempt #2. Followed Protocol Protocol 5.6: Spot Test.
Spot Test Attempt #2 with Jonathan and Brandi E.I. sample marked locations.
* Spot Test Attempt #1 with no visible plaque formations
September 18th, 2017
Spot Test Attempt #2 was discarded by Dr. Edwards due to bacterial growth. Instructed to perform a Serial Dilution using a previous successful Direct Isolation from a classmate. Followed Protocol 5.4: Picking a Plaque and Protocol 6.2 Serial Dilutions. Attempt #1 of serial dilution was discard due to using incorrect Sample Volume from Table 5.3-1. (Was using 90 microns instead of 10 microns) Six 10-fold serial dilution performed.
September 20th, 2017
Observation over Serial Dilution Attempt #1 recorded. Instructed to repeat serial dilution using saved sample from Serial Dilution Attempt #1. Followed Protocol 6.2 Serial Dilutions.
September 25th, 2017
Serial Dilution Attempt #2 performed. Followed Protocol 6.2 Serial Dilutions. No modifications noted. Attempt #2 was successful.
September 27th, 2017
Protocol 6.3: Collecting Plate Lysates performed. Specifically steps A-C. Utilized plates 10^-2 and 10^-3. And as well stored plates with phage buffer for 24 hours at 4°C prior to collection.
October 2nd, 2017
Protocol 6.4: Spot Titer performed. No modifications to protocol noted.
October 4th, 2017
Performed step G and subsequent steps after the 24 hour incubation period. The titer was estimated to be 2.0×10^9.
October 9th, 2017
Protocol 6.5: Full Plate Titer was performed. No modifications to protocol noted.
October 11th, 2017
Full plate titer estimated to be 1.9×10^9.
October 16th, 2017
First attempt at Protocol 7.1 Making Webbed Plate from a Lysate of Known Titer.
October 19th, 2017
Performed Protocol 6.3: Collecting Plate Lysates. Let phage buffer sit on plate over the weekend.
October 23rd, 2017
Collected plate lysate and labeled tube appropriately.
October 25th, 2017
My first attempt at phage DNA extraction. Utilized QuBit outline protocol according the online manual provided for our Wizard kit. Concentration was inadequate.
October 30th, 2017
Second attempt at Protocol 9.1: Phage DNA Extraction. Utilized QuBit protocol with no modifications. I made note that I believed a low reading the first round was due to poor technique on my end. Concentration from second round of phage DNA extraction was inadequate.
November 2nd, 2017
Third attempt at Protocol 9.1: Phage DNA Extraction. Modification noted on this attempt to protocol was the addition of nuclease mix to 10 micro liters. Reading was noted to be the lowest of all three attempts thus far at 1.2.
November 6th, 2017
Fourth attempt at Protocol 9.1 Phage DNA Extraction. Addition of nuclease mix was increased to 20 micro liters. Reading noted with not much improvement and no where near required amount to continue.
November 8th, 2017
Lysate was running low, I was forced to put aside temporarily phage DNA extraction and make more webbed plates to collect phage lysate. Protocol 7.1 Making Webbed Plate from a Lysate of Known Titer performed.
November 13th, 2017
Protocol 6.3: Collecting Plate Lysates performed.
November 15th, 2017
Lysate collected and the fifth round of Protocol 9.1 Phage DNA Extraction was performed. After discussion with Dr. Pierce I emphasized the slow mixing using the resin. In hopes that the additional time would allow for further break down of the capsid. All remained steps were followed according to protocol. Phage DNA extracted was sufficient at 82.5.
November 27th, 2017
Protocol 10.1: Setting Up Restriction Enzyme Digests was performed. Modifications were stated orally to me and not documented properly. Miranda Fuentes was an accurate record of the modifications to protocol required by Dr. Edwards.
November 29th, 2017
Utilized Protocol 8.1b to prepare a TEM sample for viewing.
December 4th, 2017
First attempt at Protocol 10.2: Casting Agarose Gels and Protocol 10.3: Gel Electrophoresis of Restriction Enzyme Digests. Running of gel revealed no bands. I believe that it is due to the length of time my enzyme digests were stored.
December 5th, 2017
Repeat of Protocol 10.1 and then subsequently Protocol 10.2-10.3. Gel was successful. Although it was missing a column and there was some distortion in the stain. bands were clearly visible and countable.