Discovery of MuskMan
Title: Soil Sample Collection Data
Purpose: Collect soil samples to discover new phages
Date: September 2, 2017
1
2
3
4
Title: Direct Isolation
Purpose: To extract potential phages from soil samples
Date: September 6, 2017
Procedure
1) Prepare work area for aspetic environment
2) Extract potential phage from soil sample
a) Using a 15 mL conical tube, fill it 1/3 – 1/2 with soil sample
* I filled tube with 8.5 mL of Sample #2
b) Add 2-3 mL of enrichment broth to sample
* I added 2.5 mL of enrichment broth
c) Cap tube and invert several times until mixed thoroughly
d) Incubate the tube while shaking it vigorously in a shaking incubator for 1-2 hours
* I allowed my tube to incubate for 2 hours
e) Allow the sample to sit until the particulate matter has settled to the bottom
3) Collect phage filtrate
a) Open the package of a 0.22 µm syringe filter but leave the filter in the packaging
b) Remove 2 mL of top liquid from the sample #2 tube used in step 2
c) Attach the syringe to the filter and removie it from the package
1) Be sure the filter and syringe are securely tightened
d) Using the syringe with the filter, dispense 0.5 mL of filtrate into a labeled microcentrifuge tube
4) Discard syringe and filter
5) Proceed to performing a Plaque Assay
Title: Enriched Isolation (Day 1)
Purpose: To amplify the potential phages in soil sample
Date: September 6, 2017
Procedure
1) Obtain a soil sample
* I used soil sample #1
2) Extract potential phages from that soil sample
a) Fill 15 mL of soil sample into a 50 mL conical tube
b) Add enrichment broth to the tube until it reaches the 35 mL mark
c) Vortex for 30 seconds
d) Shake the sample tube at 250 rpm for 1-2 hours
*I allowed my tube to shake for 2 hours
e) Centrifuge tube at 2000x for 10 minutes to pellet the soil
3) Prepare work area for aspetic environment
4) Filter the supernatant through a 0.22 µm filter by using a syringe. This will remove any remaining bacteria and soil particles.
a) Collect filtrate in a 50 mL conical tube
* I was able to collect roughly 20 mL of filtrate
5) Add 0.5 mL of host bacteria to the conical tube
6) Incubate the conical tube at 37°C while shaking at 220 rpm for 2-5 days.
a) Be sure to screw the cap on 1/4 of a turn so that it is loosely capped and secure it with a piece of tape so it doesn’t fall off. This will allow for proper aeration of culture mixture.
Title: Plaque Assay (Day 1)
Purpose: To determine if soil samples containned any phages
Date: September 6, 2017
Procedure
1) Prepare work area for aspetic environment
2) Gather phage samples
3) Inoculate host bacteria (M. smegmatis) with phage samples
a) Obtain the same number of aliquots of 250 µL host bacterial cultures as phage samples as well as two more for a negative and positive control
*I did not use a positive control in my plaque assay
1) label tubes accordingly
b) Dispense 10 µL of enriched phage sample into the appropriate culture tube
c) Dispense 500 µL of direct isolation phage sample into the appropriate culture tube
d) Dispense 10 µL of phage buffer into the appropriate culture tube. This will be your positive control
4) Mix each inoculated tube by tapping on it, making sure the sample and bacteria make contact.
5) Let your sample tube sit undisturbed for 5-10 minutes to allow for attachment of phage sample and bacteria
6) Plate your sample tubes with top agar
a) Label as many agar plates as you have samples
*In this scenario, their will be two plates. One for the direct and one for the negative control.
b) Remove a bottle of top agar from a 55° C bath
c) Use a sterile 5 mL pipette to aspetically transfer 3 mL of top agar to an inoculated host tube
d) Immediately aspirate the mixture from the tube back into the pipette and transfer it to the correctly labeled plate. Discard the pipette tip each time.
e) Quickly tilt the agar plate in different directions until the top agar evenly coats the the plate.
f) Repeat steps (c), (d), and (e) for each inoculated host tube sample
7) Allow plates to sit for 20 minutes so they can solidify
8) Invert plates and place in incubator at 37° C for 24-48 hours.
* I incubated my plates at 1:25 PM
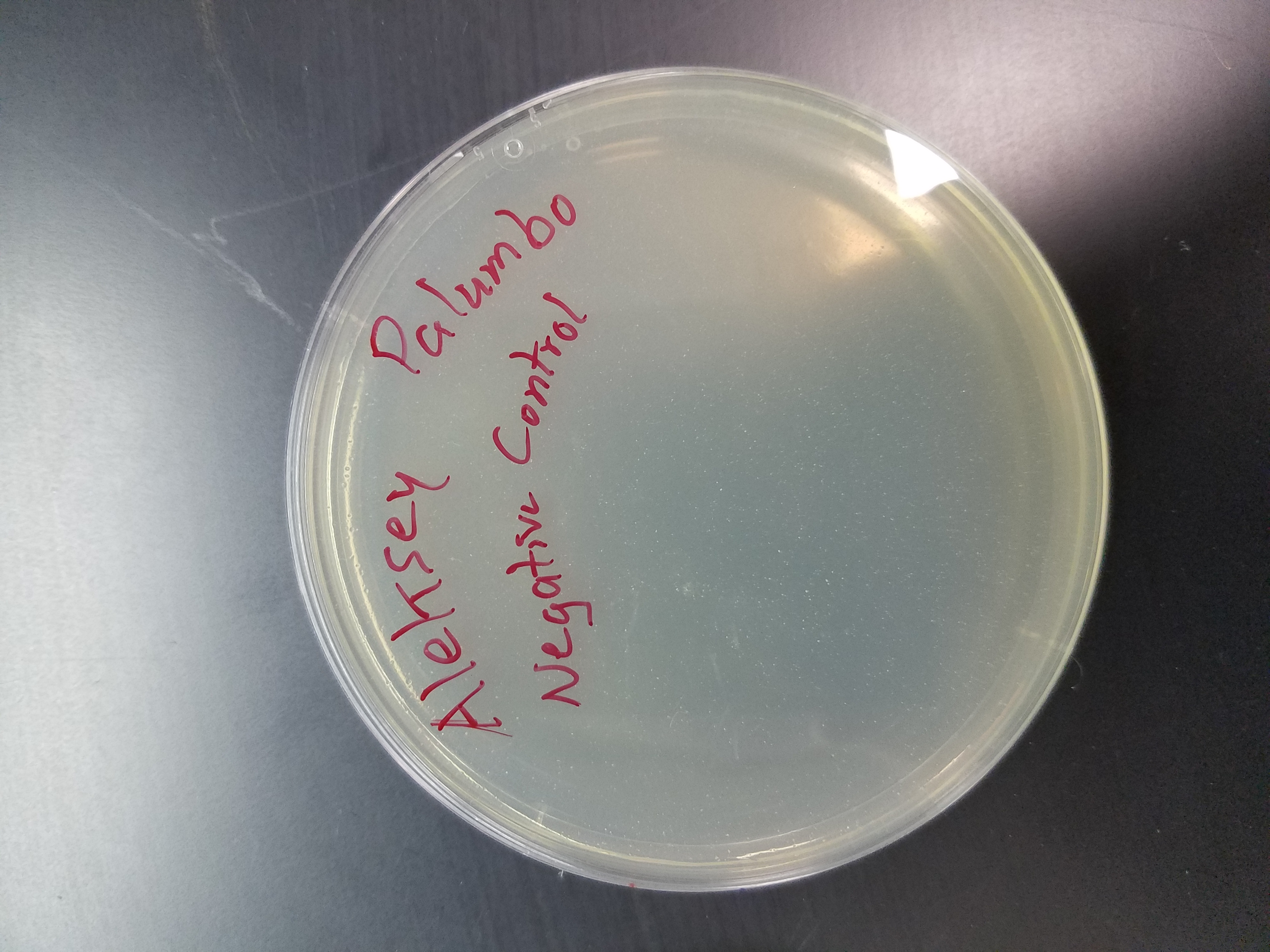
Title: Plaque Assay (Day 2)
Purpose: To determine if soil samples containned any phages
Date: September 8, 2017
Procedure
1) Check agar plates for any signs of plaques
a) If no plaques are visible, return plates to incubator and check again later on
2) Record observations in lab journal
* I did not see any signs of plaques on any of my plates.

Title: Enriched Isolation (Day 2)
Purpose: To amplify the potential phages in soil sample
Date: September 11, 2017
Procedure
1) Prepare work area for aspetic environment
2) Obtain enriched culture sample
3) Transfer 1.4 mL of enriched sample to a microcentrifuge tube
a) Repeat this step so that you have two microcentrifuge tubes, each containing 1.4 mL of enriched culture
4) Centrifuge the two microcentrifuge tubes for 1 minute to pellet the bacteria
5) If you think your enrichment contains any non-host bacteria, filter the supernatant.
a) Remove the plunger from a syringe
b) Attach a sterile 0.22 µm filter to the barrel of the syringe
c) Pipette 1 mL of supernatant out of each microcentrifuge tube into the barrel of the syringe
d) Insert the plunger into the syringe barrel and depress the filtrate into a new sterile microcentrifuge tube
6) Label the microcentrifuge tube accordingly and store it at 4°C.
7) Proceed to perform a Spot Test with the enriched culture filtrate.
Title: Spot Test (Trial #1)
Purpose: To test a sample for the presence of any phages
Date: September 11, 2017
Procedure
1) Prepare work area for aspetic environment
2) Gather phage culture samples to test
* My direct and enriched showed no signs of plaques on my plaque assay from before so I will only test my enriched culture sample.
3) Prepare a bacterial lawn
a) Label the bottom of an agar plate and divide it into as many sections as you have samples
*I labeled my plate with a 3×3 grid
b) Collect a 250 µL culture tube of host bacteria and 3 mL molten top agar
c) Use a 5 mL sterile pipette to transfer 3 mL of molten top agar to a 250 µL culture tube containing host bacteria
d) Quickly pipette up the solution and dispense it onto the labeled agar plate
1) Be sure to tilt the plate in multiple directions until the plate is evenly coated in the molten agar mixture
e) Let the plate sit undisturbed for 20 minutes to allow the top agar to solidify
4) Spot the phage culture samples and controls on the top agar lawn
a) Aseptically transfer 10 µL of each sample onto the correct location on the agar plate as depicted by the drawn grid
* I spotted three of the grid boxes with the enriched culture sample
b) Transfer 10 µL of sterial phage buffer on the plate as a negative control
c) Wait 10-15 minutes to allow the liquid from the samples to absorb into the agar
d) Without inverting the plates, incubate them at 37° C for 24-48 hours
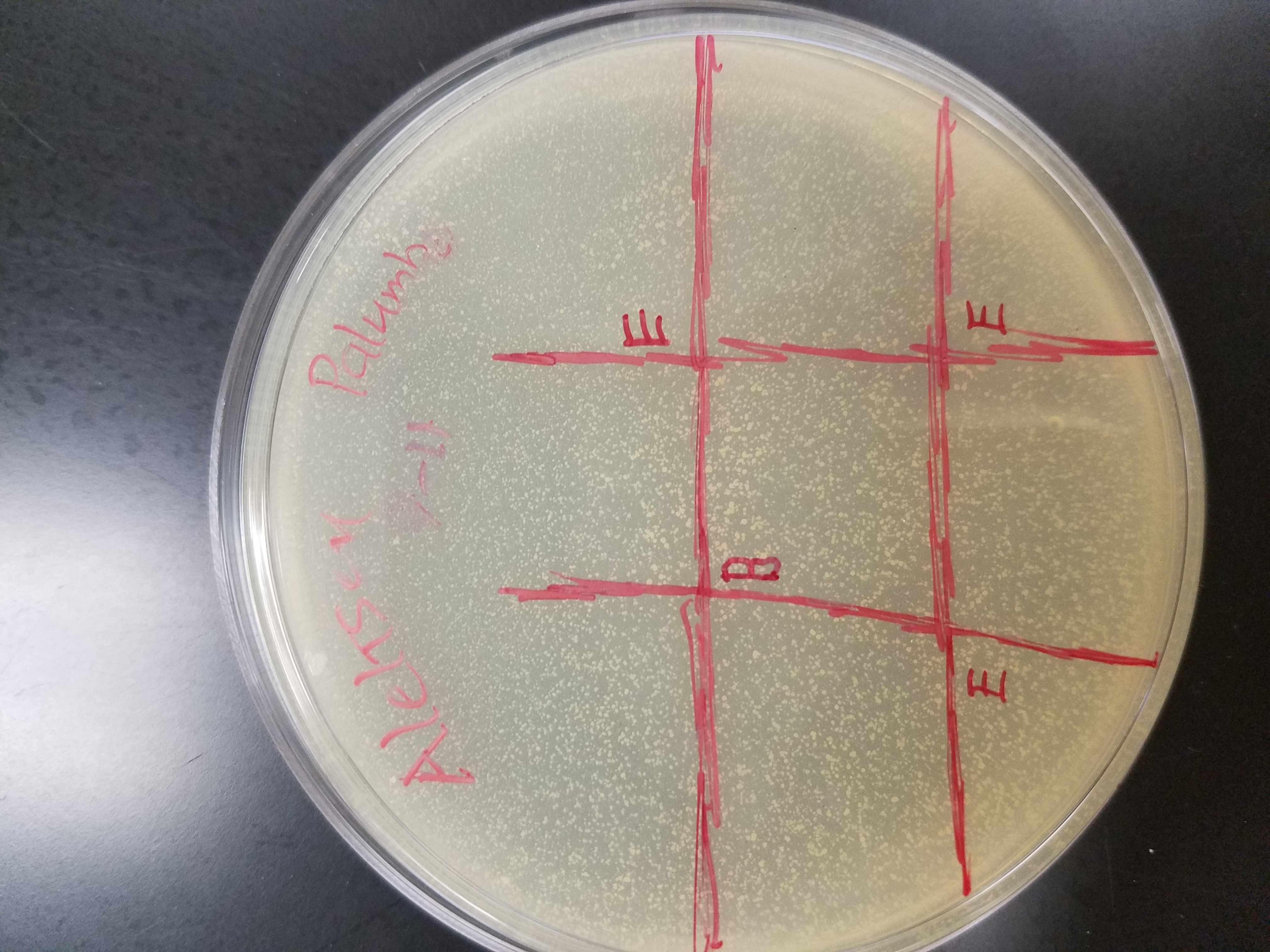
Title: Spot Test (Trial #1)
Purpose: To test a sample for the presence of any phages
Date: September 13, 2017
Procedure
1) Check plate for any sign of clearing
* My agar plate had no signs of clearing/spotting, indicating that my sample didn’t contain any phages.
Title: Spot Test (Trial #2)
Purpose: To test a sample for the presence of any phages
Date: September 13, 2017
Procedure
1) Prepare work area for aspetic environment
2) Gather phage culture samples to test
* My previous spot test came up negative in terms of plaques so I adopted Brittaney’s enriched culture sample to test with
*Brittany’s enriched soil sample was collected on Sept. 6, 2017 at 32°13’22″N 98°11’55″W
3) Prepare a bacterial lawn
a) Label the bottom of an agar plate and divide it into as many sections as you have samples
*I labeled my plate with a 3×3 grid
b) Collect a 250 µL culture tube of host bacteria and 3 mL molten top agar
c) Use a 5 mL sterile pipette to transfer 3 mL of molten top agar to a 250 µL culture tube containing host bacteria
d) Quickly pipette up the solution and dispense it onto the labeled agar plate
1) Be sure to tilt the plate in multiple directions until the plate is evenly coated in the molten agar mixture
e) Let the plate sit undisturbed for 20 minutes to allow the top agar to solidify
4) Spot the phage culture samples and controls on the top agar lawn
a) Aseptically transfer 10 µL of each sample onto the correct location on the agar plate as depicted by the drawn grid
* I spotted three of the grid boxes with the enriched culture sample
* I only used 5 µL of each sample to allow for a more controlled transfer
b) Transfer 10 µL of sterial phage buffer on the plate as a negative control
* I only used 5 µL to allow for a more controlled transfer
c) Wait 10-15 minutes to allow the liquid from the samples to absorb into the agar
d) Without inverting the plates, incubate them at 37° C for 24-48 hours
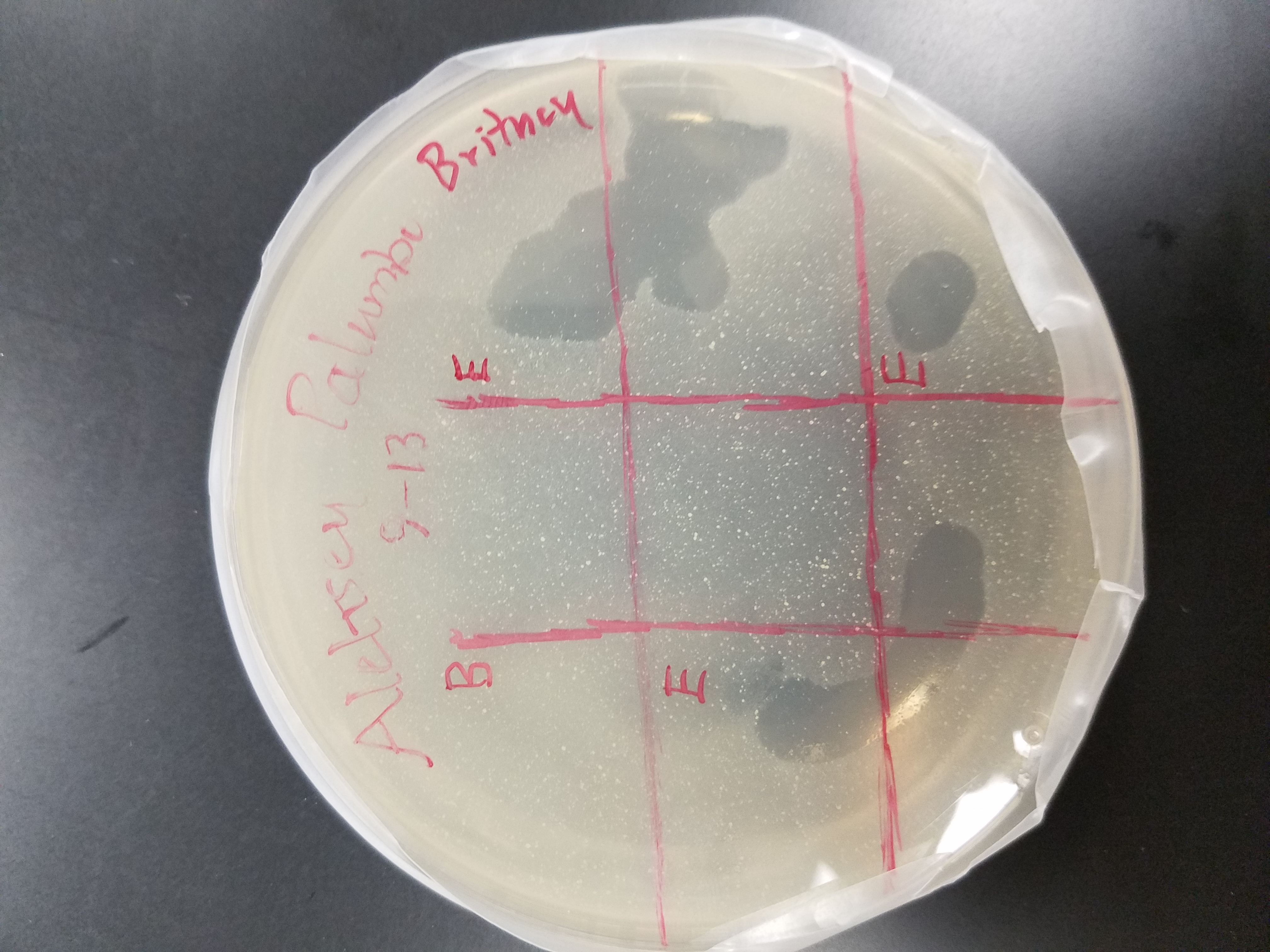
Title: Spot Test (Trial #2)
Purpose: To test a sample for the presence of any phages
Date: September 15, 2017
Procedure
1) Check plate for any sign of clearing
* My agar plate showed signs of clearing. The areas of cleared appeared quite large and were in all three coordinates labeled enriched.
Title: Serial Dilutions
Purpose: To decrease the concentration of a liquid phage sample
Date: September 20, 2017
Procedure
1) Prepare work area for aspetic environment
2) Prepare a 10-fold serial dilution
a) Label 8 microcentrifuge tubes 10^-1, 10^-2…..10^-8
* I only diluted the enriched sample up to 10^-6
*I used Britney’s enriched sample since my sample didn’t have any phages
b) Add 90 µL phage buffer to each tube
3) Perform a 10-fold serial dilution
a) Add 10 µL of phage sample to the 10^-1 tube and vortex well
b) Transfer 10 µL from the 10^-1 tube into the 10^-2 tube and vortex well
*Be certain to change pipette tips after every transfer
c) Continue to do this until you vortex the last tube
4) Perform a plaque assay for each of the diluted microcentrifuge tubes according to the previously noted procedure.
*Dispense 10 µL of each diluted phage tube into the host bacteria tube
a) Allow plates to incubate at 37°C for 24-48 hours
Title: Serial Dilution Results
Purpose: To determine if a plaque assay can be isolated from
Date: September 22, 2017
Procedure
1) See if any of the diluted plaque assays displayed any plaques
2) Note these appearance in your laboratory journal
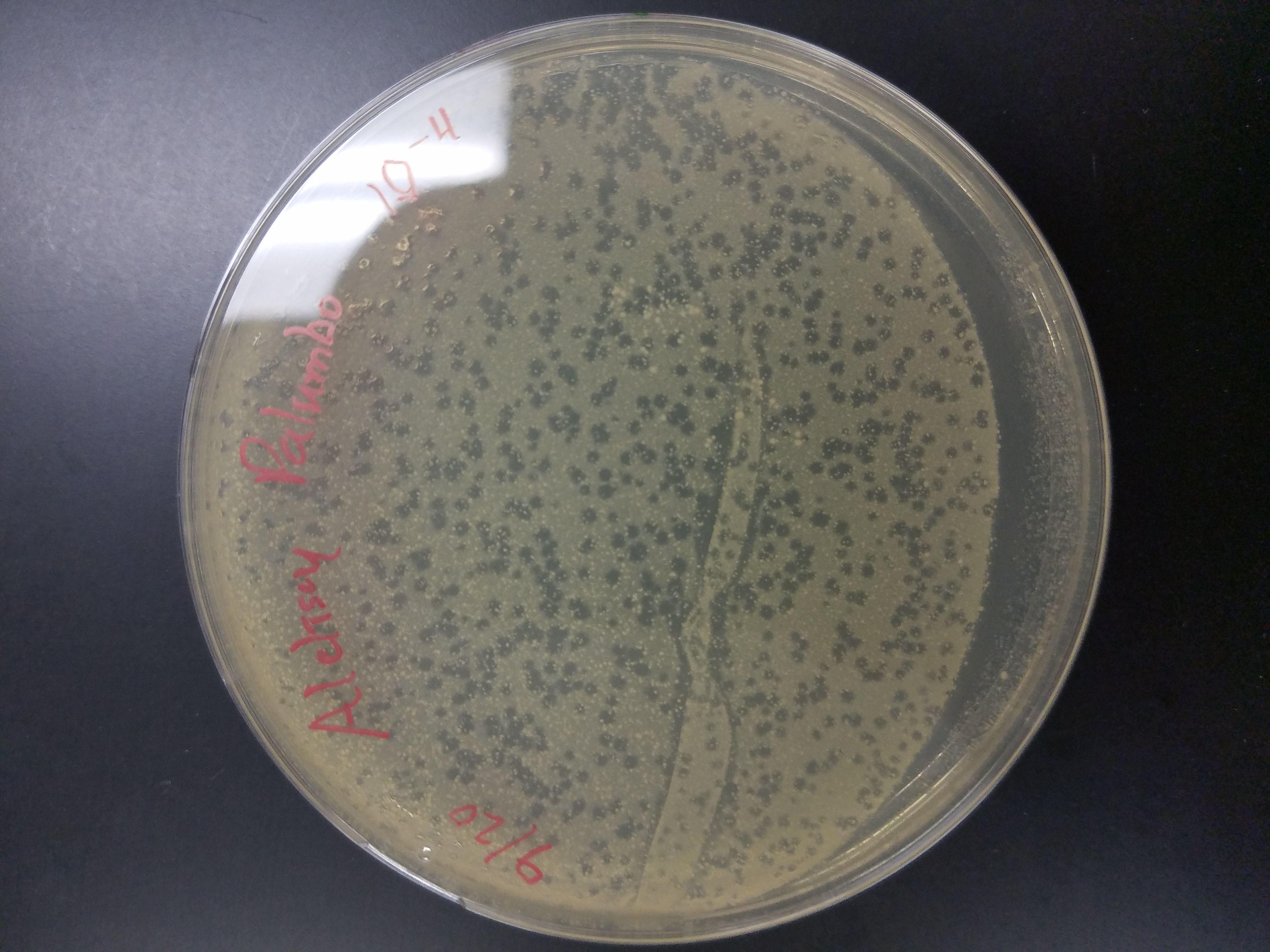
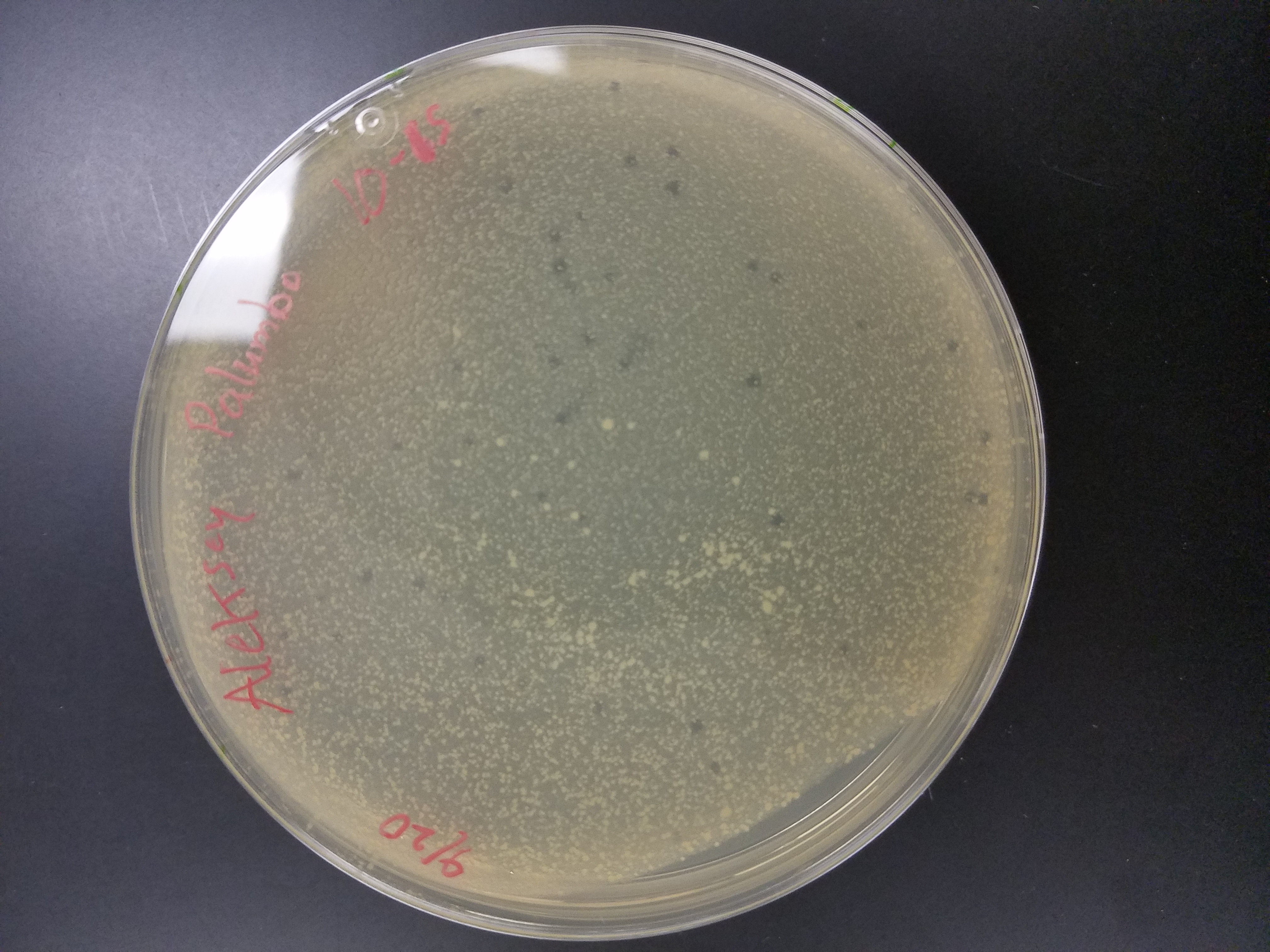
*10^-4 plate had over 100 plaques
*10^-5 plate had roughly 15 plaques
Title: Making Webbed Plates from a known Titer
Date: October 16, 2017
Purpose:
I will attempt to create several webbed plates from my known titer in order to create more lysate for further experiments
Title: Switching Phages
Date: November 1, 2017
After several failed attempts at trying to make webbed plates from my high titer lysate, I have decided to partner with Amanda Stone and help her isolate her phage, Noonan. I feel this will benefit us both in learning about bacteriophages and laboratory techniques throughout the class.
Title: DNA Extraction attempt #1
Date: November 8, 2017
Purpose: To obtain pure DNA from our isolated bacteriophage
Amanda and I attempted to isolate the DNA from our bacteriophage by performing a certain procedure. Unfortunatley, we were only able to extract DNA that had a concentration of 23 ng/µL, which is not enough to be sequenced.
Title: DNA Extraction attempt #2
Date: November 15, 2017
Purpose: To obtain pure DNA from our isolated bacteriophage
Amanda and I attempted to isolate the DNA from our bacteriophage by performing a certain procedure. Unfortunatley, we were only able to extract DNA that had a concentration of 46 ng/µL, which is not enough to be sequenced.
Title: DNA Extraction attempt #3
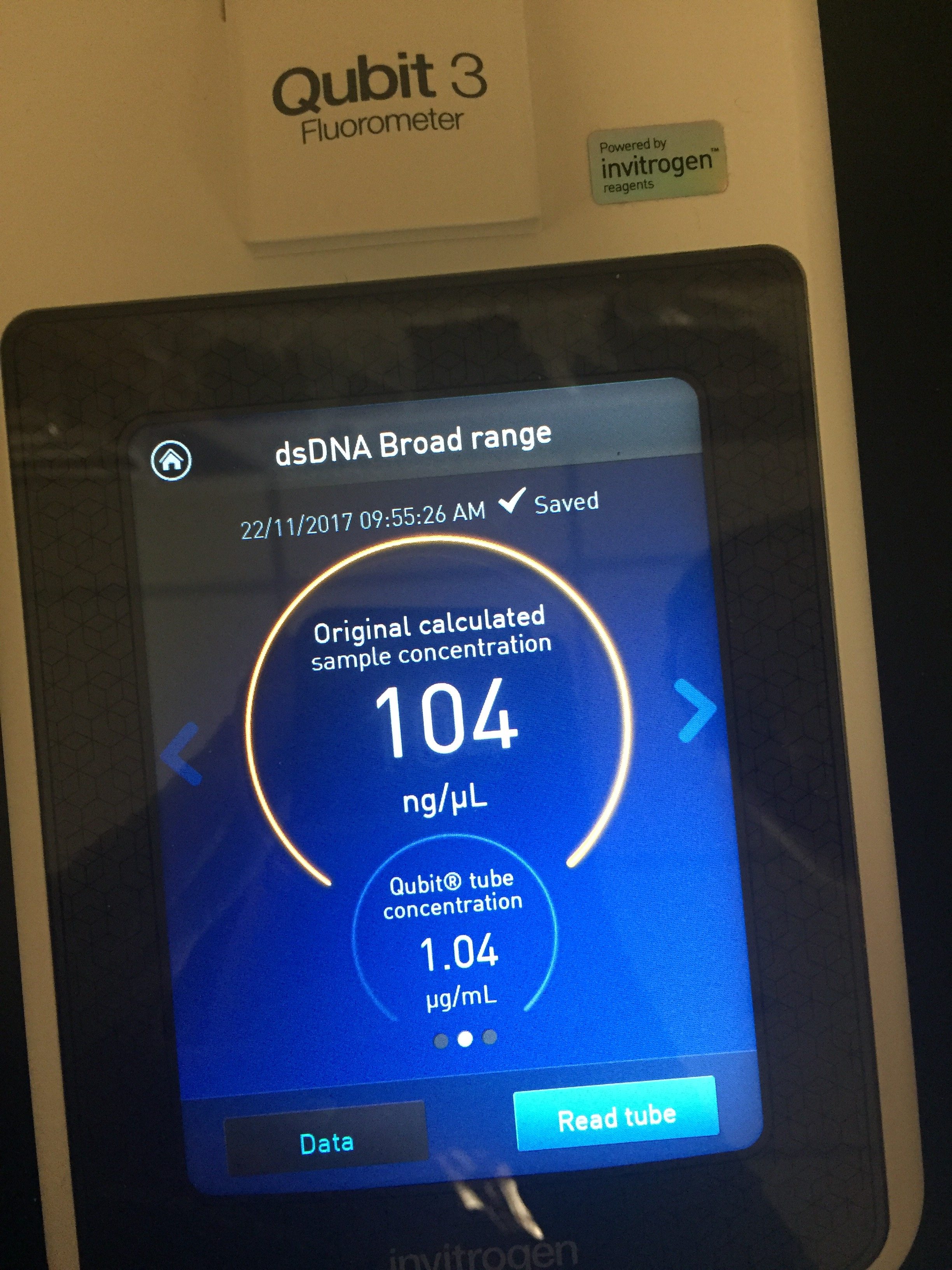
Date: November 20, 2017
Purpose: To obtain pure DNA from our isolated bacteriophage
Amanda and I attempted to isolate the DNA from our bacteriophage by performing a certain procedure. We were able to extract DNA that had a concentration of 104 ng/µL, which is enough to be sequenced.
Title: Bacteriophage Imaging
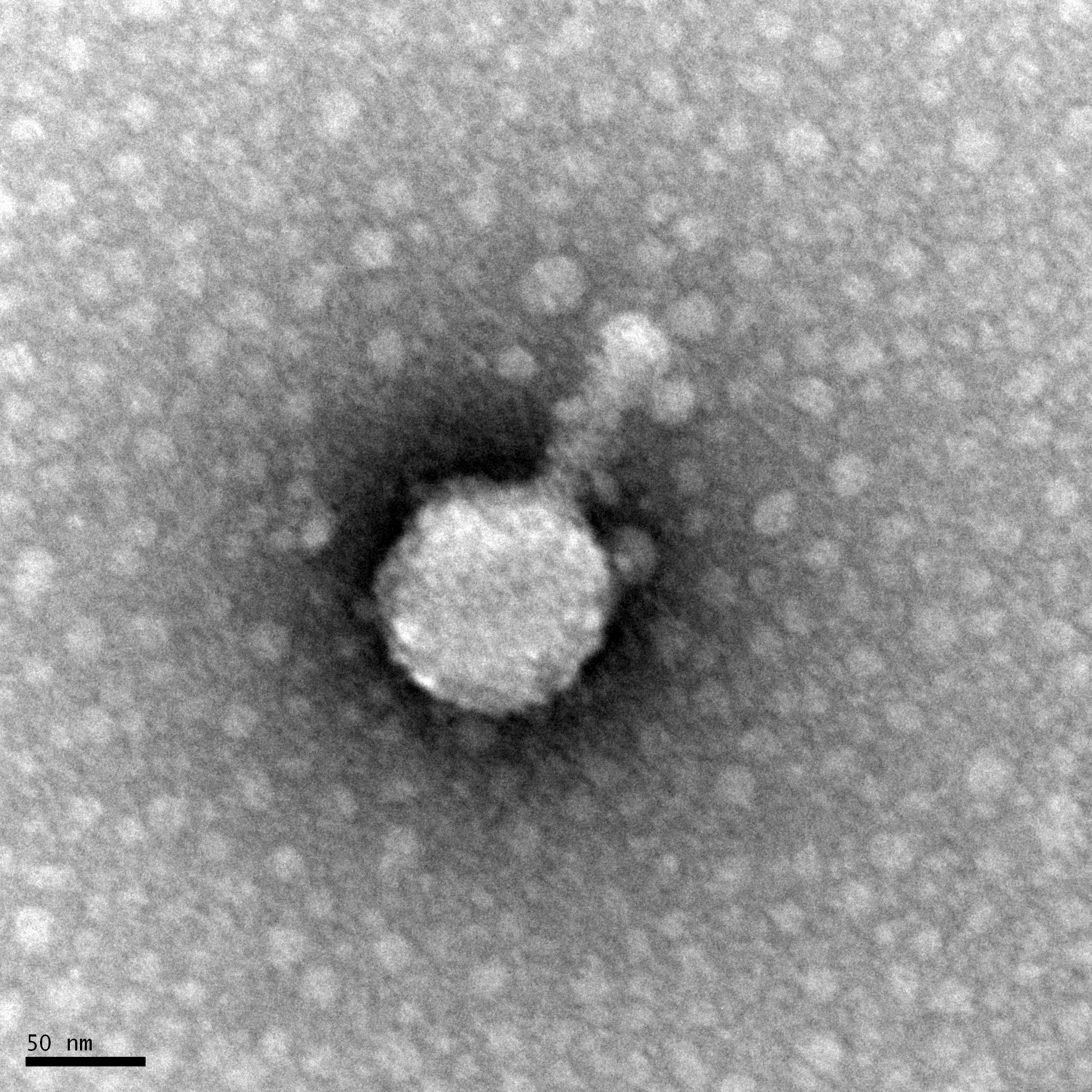
Date: November 27, 2017
Purpose: To obtain a visual image of our isolated bacteriophage, Noonan.
Amanda and I followed a procedure to be able to visual our bacteriophage, Noonan. We used the technology of Transmission Electron Microscopy (TEM) to do this. We first mounted our isolated bacteriophage onto a grid so it can be visualized under an electon microscope in a different laboratory.
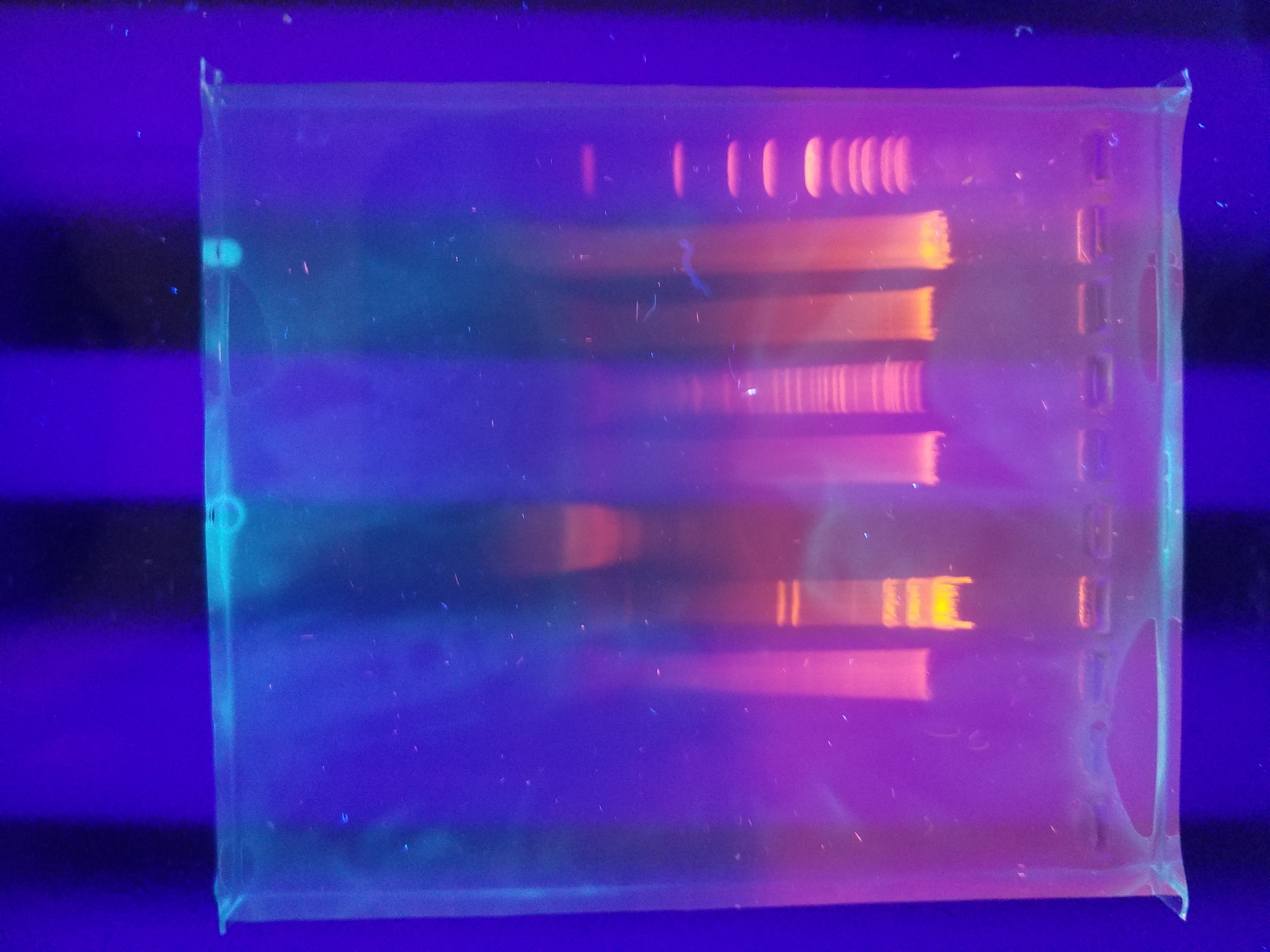
Title: Phage Genome Analysis using Restriction Enzyme
Date: December 6, 2017
Purpose: To further characterize Noonan as a bacteriophage.
Amanda and I used a variety of restriction enzymes to be able to genetically characterize our bacteriophage, Noonan. We used the follwing restriction enzymes: BamHI, CIaI, EcoRI, HaeIII, HindIII, and SaII. We were able to successfully do this by running our bacteriophage’s DNA through a gel electrophoresis. Below is a picture of our results.