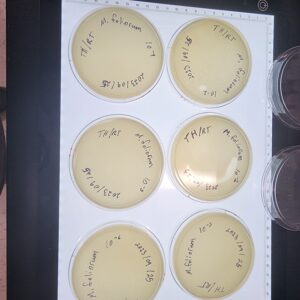
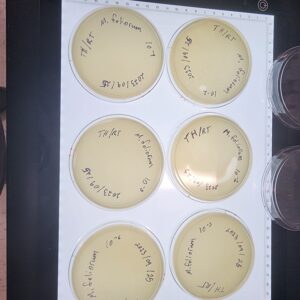
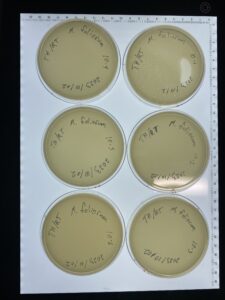
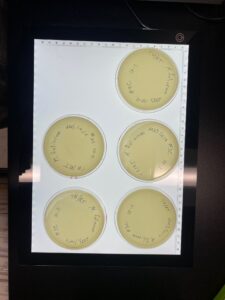
Discovery of Tarleton
Tarleton Information
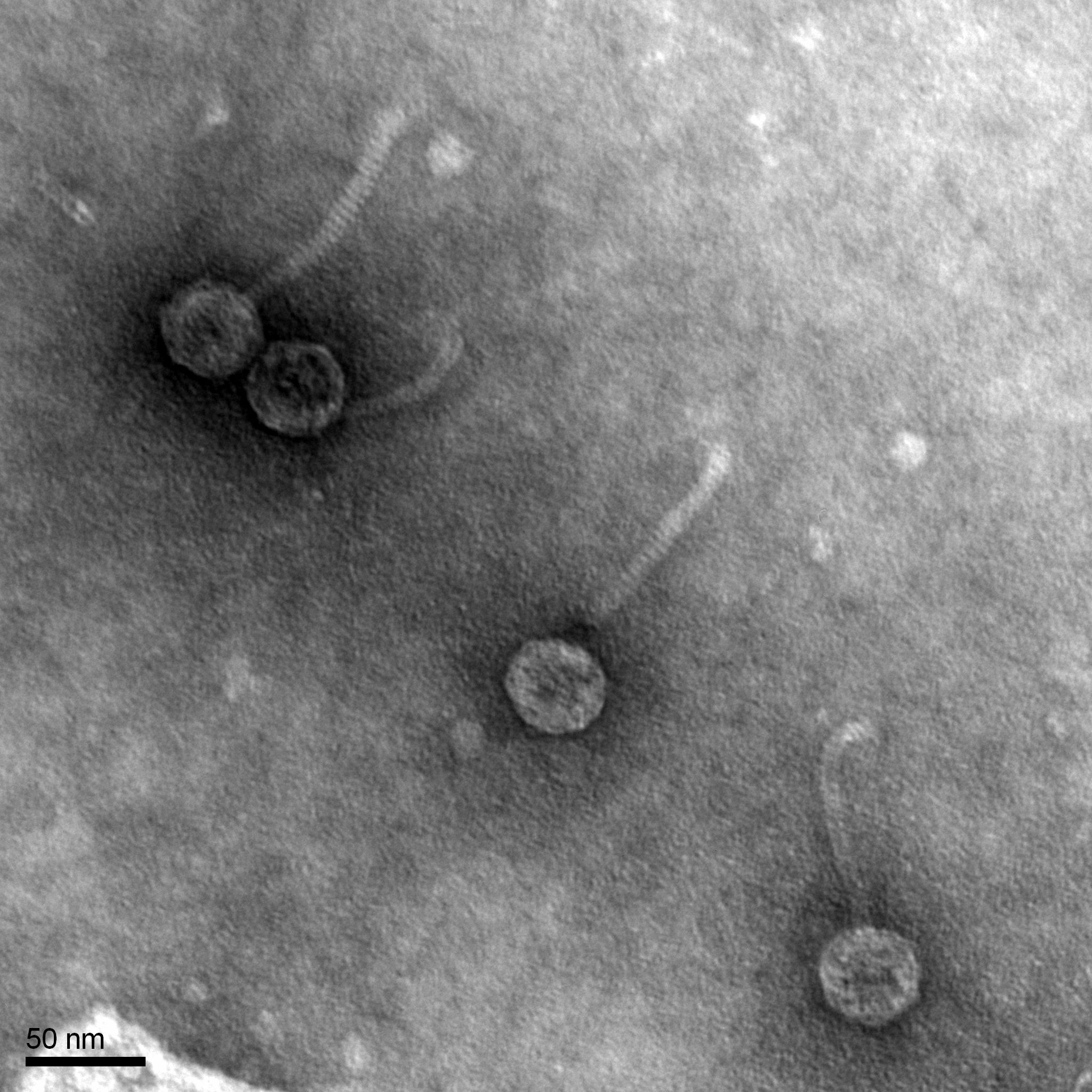
Morphology: Siphoviridae
Sample Collection
| Collector Name |
Ricardo Torres | Tim Hester | Tim Hester | Tim Hester | Ricardo Torres |
| Sample No. | 1 | 2 | 3 | 4 | 5 |
| Date of Collection | 08/28/2020 | 08/28/2020 | 09/05/2023 | 09/05/2023 | 09/13/2023 |
| Sample Type | Soil | Soil | Soil | Soil | Soil |
| General Location | Bosque river trail | Dog Park | Forest Floor | Prairie | Storm Drain Outside the Science Building |
| Location Description | Creek bank | Dog Park behind apt. | Forest floor in Erath county | Prairie in Erath county | Tarleton Stephenville Campus |
| GPS Coordinates | 32.217062, -98.193983 | 32.2190767, -98,2161289 | 32.103299, -98.3874912 | 32.1028864, -98.3873759 | 32.21667, -98.21992 |
| Sample Depth | Surface | 2 inches under the surface | Surface Layer | Surface layer | Surface Layer |
| Ambient Temperature | 35.5°C | 35°C | 40.5°C | 40.5 | 26.7 |
Sample Collection
| Collector Name |
Tim Hester | Ricardo Torres | #### | #### | #### |
| Sample No. | 6 | 7 | Adoption | ## | ## |
| Date of Collection | 09/13/2023 | 09/20/2023 | 09/17/2023 | ##/##/#### | ##/##/#### |
| Sample Type | Soil | Soil | Soil | #### | #### |
| General Location | Southwest of the science building | N. Lillian st. southwest of fine arts buildng |
2732 Clovermeadow Dr Fort Worth Tx 76123 |
#### | #### |
| Location Description | Under a tree | Under a rose bush | Sidewalk near house | #### | #### |
| GPS Coordinates | 32.2168529, -98.2204037 | 32.21618, -98,21837 | #### | #### | #### |
| Sample Depth | Surface | Surface | Surface | #### | #### |
| Ambient Temperature | 26.7°C | 35.6°C | 27.8°C | #### | #### |
Isolation/Purification
Title: Isolation/Purification of Environmental Sample by Direct Isolation and Plaque Assay
Date: 23/08/30
Redo: No
Sample: 1
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- Initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added PYCa media using the 5 mL serological pipette to the sample in a 15mL conical tube and inverted until the media was thoroughly mixed with the sample.
- It was then left in a rotating incubator for one hours at 220 rpm 29 degrees celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe and .22 um syringe filter we then removed the liquid sample from the 15 mL conical tube and transferred over the now filtered sample into a microcentrifuge tube.
Plaque assay
1. Using a 100 ul micropipette, we transferred 500 microliters of the filtered sample into a test tube containing 250 microliters of host bacteria.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes
7. Plate was then incubated for 96 hours @ 29 degrees C starting 1114 hrs. on 23/08/30.
The plate was removed from the incubator in order to check the status of the phage development at 14:03 hrs. on 09/01/2023.
Results:
Results were negative. No plaques produced. Sample 1 plate was removed from incubator and disposed of with other contaminated samples.
Conclusions and Next Steps:
Sample was contaminated, sterile, negative for phages effecting M. foliorum bacteria. New samples to be collected 9/5/2023.
Title: Isolation/Purification of Environmental Sample by Direct Isolation and Plaque Assay
Date: 23/08/30
Redo: yes
Sample: 2
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added PYCa media using the 5 mL serological pipette to the sample in a 15mL conical tube and inverted until the media was thoroughly mixed with the sample.
- It was then left in a rotating incubator for one hours at 220 rpm 29 degrees celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe and .22 um syringe filter we then removed the liquid sample from the 15 mL conical tube and transferred over the now filtered sample into a microcentrifuge tube.
Plaque assay
1. Using a 100 ul micropipette, we transferred 500 microliters of the filtered sample into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes and was then incubated for 96 hours.
7. Plate was then incubated for 96 hours @ 29 degrees C starting 1114 hrs. on 23/08/30.
The plate was removed from the incubator in order to check the status of the phage development at 1403 hrs. on 09/01/2023.
Results:
Results were negative. No plaques produced. It was noted that the sample media did not cover the whole plate.
Conclusions and Next Steps:
Sample was contaminated, sterile, or negative for phages effecting M. foliorum bacteria. New samples to be collected 9/5/2023. During the next attempt more care will be taken to assure complete coverage of the plate by the sample.
Title: Isolation/Purification of Environmental Sample by Enriched isolation and Plaque Assay
Date: 23/09/11
Redo: yes
Sample: 3
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added 15 mL of sample to a 50 mL conical tube .
- Added PYCa media to sample up to the 35 mL mark on the conical tube.
- Sample was then left in a rotating incubator for one hours at 220 rpm at 29 degrees Celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe the supernatant was aspirated from the conical tube and placed in a vacuum filtration apparatus with a 22 um filter. 7.
- The sample was then aspirated through the filter into a 50 mL conical tube.
- 0.5 mL of M foliorum bacterial sample was then added to the sample in conical tube.
- The sample was then placed in a shaker at 220 rpm for 5 days. To assure the sample remains aerated the lid for the conical tube was unscrewed 1/4 turn and secured in place with tape.
- After 5 days incubation lab bench work station was sterilized again using aseptic techniques.
- 2 micro centrifuge tubes were obtained and 1.4 mL of enriched culture was added to each micro tube from the 50 mL conical tube containing the sample using a 2mL pipette.
- The 2 micro centrifuge tubes were then placed in the centrifuge on high speed for 1 minute to pellet the bacteria.
- As the sample was still cloudy a syringe and 0.22 um filter were used to clarify the sample.
- The plunger was removed from the syringe and 1mL of sample was removed from each tube leaving the pellet in the tube and added to the open syringe.
- The tip of the filter was then placed into a new micro centrifuge tube and the plunger inserted back into the syringe to dispense the 2mL of filtered sample into the new tube.
- The tube was then capped to await plaque assay.
- Plaque assay
- 1. Using a 10 ul micropipette, we transferred 10 microliters of the filtered enriched sample into a test tube containing 250 microliters of host bacteria.
-
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes.
7. Plate was then incubated for 48 hours @ 29 degrees C starting 1114 hrs. on 23/09/11.
The plate was removed from the incubator in order to check the status of the phage development at 1403 hrs. on 09/12/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was contaminated, sterile, negative for phages effecting M. foliorum bacteria. New samples to be collected 9/5/2023.
Title: Isolation/Purification of Environmental Sample by Direct Isolation and Plaque Assay
Date: 23/09/11
Redo: yes
Sample: 4
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added PYCa media using the 5 mL serological pipette to the sample in a 15mL conical tube and inverted until the media was thoroughly mixed with the sample.
- It was then left in a rotating incubator for one hours at 220 rpm 29 degrees celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe and .22 um syringe filter we then removed the liquid sample from the 15 mL conical tube and transferred over the now filtered sample into a microcentrifuge tube.
Plaque assay
1. Using a 100 ul micropipette, we transferred 500 microliters of the filtered sample into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes.
7. Plate was then incubated for 48 hours @ 29 degrees C starting 1114 hrs. on 23/09/11.
The plate was removed from the incubator in order to check the status of the phage development at 1403 hrs. on 09/12/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. New samples to be collected 9/13/2023.
Title: Isolation/Purification of Environmental Sample by Enriched isolation and Plaque Assay
Date: 23/09/14
Redo: yes
Sample: 5
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added 15 mL of sample to a 50 mL conical tube .
- Added PYCa media to sample up to the 35 mL mark on the conical tube.
- Sample was then left in a rotating incubator for one hours at 220 rpm at 29 degrees Celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe the supernatant was aspirated from the conical tube and placed in a vacuum filtration apparatus with a 22 um filter. 7.
- The sample was then aspirated through the filter into a 50 mL conical tube.
- 0.5 mL of M foliorum bacterial sample was then added to the sample in conical tube.
- The sample was then placed in a shaker at 220 rpm for 5 days. To assure the sample remains aerated the lid for the conical tube was unscrewed 1/4 turn and secured in place with tape.
Plaque assay
- Using a 10 ul micropipette, we transferred 10 microliters of the filtered enriched sample into a test tube containing 250 microliters of host bacteria.
- The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes.
7. Plate was then incubated for 48 hours @ 29 degrees C starting 0957 hrs. on 23/09/18. 8. Plate was removed from the incubator in order to check the status of the phage development at 1450 hrs. on 09/19/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. Sample 6 collected 9/13/2023 to be used.
Title: Isolation/Purification of Environmental Sample by Direct Isolation and Plaque Assay
Date: 23/09/18
Redo: yes
Sample: 6
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added PYCa media using the 5 mL serological pipette to the sample in a 15mL conical tube and inverted until the media was thoroughly mixed with the sample.
- It was then left in a rotating incubator for one hours at 220 rpm 29 degrees celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe and .22 um syringe filter we then removed the liquid sample from the 15 mL conical tube and transferred over the now filtered sample into a microcentrifuge tube.
Plaque assay
1. Using a 100 ul micropipette, we transferred 500 microliters of the filtered sample into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes.
7. Plate was then incubated for 48 hours @ 29 degrees C starting 1039 hrs. on 23/09/18. 8. Plate was removed from the incubator in order to check the status of the phage development at 1450 hrs. on 09/19/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. New samples to be collected 9/20/2023.
Title: Isolation/Purification of Environmental Sample by Direct Isolation and Plaque Assay
Date: 23/09/20
Redo: yes
Sample: 7
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- We added PYCa media using the 5 mL serological pipette to the sample in a 15mL conical tube and inverted until the media was thoroughly mixed with the sample.
- It was then left in a rotating incubator for one hours at 220 rpm 29 degrees celsius.
- The sample was then centrifuged at 2000 gravities for 10 minutes.
- Using a 5mL syringe and .22 um syringe filter we then removed the liquid sample from the 15 mL conical tube and transferred over the now filtered sample into a microcentrifuge tube.
Plaque assay
1. Using a 100 ul micropipette, we transferred 500 microliters of the filtered sample into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
6. The plate was then allowed to solidify for 20 minutes.
7. Plate was then incubated for 48 hours @ 29 degrees C starting 1039 hrs. on 23/09/18. 8. Plate was removed from the incubator in order to check the status of the phage development at 1450 hrs. on 09/19/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. Adoption of Phage from another group recommended.
Title: Isolation/Purification of Environmental Sample by Serial Dilution, and Plaque Assay
Date: 23/09/25
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-1 to 10-6.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick a single plaque to study. Circle and label plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing.
- Switch to the 10uL pipettor and remove 10uL from tube 10-1 and add it to tube 10-2 then vortex tube 10-2.
- Repeat step six until you get to the 10-6 dilution.
- Seal Plaque sample plate and return to the isolation bin in the refrigerator.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-1 to 10-6.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1039 hrs. on 23/09/25
-
9. Plate was removed from the incubator in order to check the status of the phage development at 1450 hrs. on 09/26/2023.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. When picking the plaque we may not have gone deep enough for the phage to be present. Repeat picking a plaque procedure and plaque assay.
Title: Isolation/Purification of Environmental Sample by Serial Dilution, and Plaque Assay
Date: 23/09/27
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-1 to 10-6.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick a single plaque to study. Circle and label plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing.
- Switch to the 10uL pipettor and remove 10uL from tube 10-1 and add it to tube 10-2 then vortex tube 10-2.
- Repeat step six until you get to the 10-6 dilution.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-1 to 10-6.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1039 hrs. on 23/09/27
9. Plate was removed from the incubator in order to check the status of the phage development at 1450 hrs. on 09/28/2023.
- 23/09/29 Plates removed from incubator and placed in refrigerator for examination on Monday 23/10/02.
Results:
Results were negative. No plaques produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. When picking the plaque we may not have gone deep enough for the phage to be present. Repeat picking a plaque procedure and plaque assay.
Title: Isolation/Purification of Environmental Sample by Serial Dilution, and Plaque Assay
Date: 23/10/02
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-1 to 10-6.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick a single plaque to study. Circle and label plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing.
- Switch to the 10uL pipettor and remove 10uL from tube 10-1 and add it to tube 10-2 then vortex tube 10-2.
- Repeat step six until you get to the 10-6 dilution.
- Seal Plaque sample plate and return to the isolation bin in the refrigerator.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-1 to 10-6.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1028 hrs. on 23/10/02.
-
9. Plate was removed from the incubator in order to check the status of the phage development at 1402 hrs. on 10/03/2023.
10. 23/10/03 Plates removed from incubator and placed in refrigerator for examination on Monday 23/10/04.
Results:
Results were positive. Plaques were produced on plates 10-1, 10-2, and 10-3.
Conclusions and Next Steps:
Sample was positive for phages effecting M. foliorum bacteria. Further isolation to begin immediately with a second serial dilution.
Amplification
Title: Amplification of Environmental Sample by Second Serial Dilution, and Plaque Assay
Date: 23/10/04
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-1 to 10-6.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick a single plaque to study. Circle and label plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing.
- Switch to the 10uL pipettor and remove 10uL from tube 10-1 and add it to tube 10-2 then vortex tube 10-2.
- Repeat step six until you get to the 10-6 dilution.
- Seal Plaque sample plate and return to the isolation bin in the refrigerator.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-1 to 10-6.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1028 hrs. on 23/10/04.
-
9. Plate was removed from the incubator in order to check the status of the phage development at 1145 hrs. on 10/05/2023.
10. 23/10/06 Plates removed from incubator and placed in refrigerator for examination on Monday 23/10/09.
Results:
Results were negative. No plaques were produced.
Conclusions and Next Steps:
Sample was negative for phages effecting M. foliorum bacteria. Further isolation to begin immediately with a second serial dilution. When picking the plaque we may chosen a viable plaque with the phage present. Repeat picking a plaque procedure and plaque assay.
Title: Amplification of Environmental Sample by Second Serial Dilution, and Plaque Assay
Date: 23/10/09
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-1 to 10-6.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick a single plaque to study. Circle and label plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing.
- Switch to the 10uL pipettor and remove 10uL from tube 10-1 and add it to tube 10-2 then vortex tube 10-2.
- Repeat step six until you get to the 10-6 dilution.
- Seal Plaque sample plate and return to the isolation bin in the refrigerator.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-1 to 10-6.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1028 hrs. on 23/10/09.
-
9. Plate was removed from the incubator in order to check the status of the phage development at 1535 hrs. on 10/10/2023.
Results:
Results were positive. Plaques were produced on plates 10-1 and 10-2.
Conclusions and Next Steps:
Sample was positive for phages effecting M. foliorum bacteria but in insufficient numbers to go to the next step. Further isolation to begin immediately with a second serial dilution. When picking the plaque we may chosen a viable plaque with the phage concentrations. Repeat picking a plaque procedure and plaque assay.
Title: Amplification of Environmental Sample by Second Serial Dilution, and Plaque Assay
Date: 23/10/11
Redo: yes
Sample: Adoption
Purpose: To isolate bacteria phages from 3 different plaques from an environmental sample.
Notes:
- initiate aseptic technique procedures. Wash hands and then sterilize the bench with CiDecon and 70% ethanol. After sterilizing remove gloves and light bunsen burner to maintain the sterile environment.
- Collected and labeled six micro tubes 10-0 and 10-1 for 3 different plaque samples.
- Using a p200 pipettor add 90 uL of phage buffer solution to each micro tube.
- Remove phage sample plate from refrigerator, and pick three isolated plaques to study. Circle and label each plaque on sample plate.
- Using a p200 micro pipettor gently pierce the top layer of auger in the center of the first selected plaque and submerge the tip in the first micro tube, tapping the sides, then vortex the tube to assure thorough mixing. This is tube 10-0.
- Switch to the 10uL pipettor and remove 10uL from tube 10-0 and add it to tube 10-1 then vortex tube 10-1.
- Repeat steps five and six until you get to the 10-1 dilution for all 3 plaques.
- Seal Plaque sample plate and return to the isolation bin in the refrigerator.
Plaque assay
1. Using a 10 ul micropipette, we transferred 10 microliters of the serial dilution into a test tube containing 250 microliters of host bacteria M foliorum.
2. The tube was then tilted to allow for a proper mixture between the sample and bacteria and allowed to sit for 10 minutes.
3. An agar plate was removed from the fridge and labeled to allow for quick transfer of upcoming steps.
4. After we allowed the mixture to sit for 10 minutes, using a 5 mL serological pipette we transferred 3 mL of molten top agar into the test tube, and then immediately after we aspirated it back into the syringe.
5. The mixture now containing molten top agar was then dispensed onto the agar plate and tilted to allow the liquid to completely cover the surface of the agar.
-
6. Repeat steps 1-5 for each sample dilution 10-0 to 10-1.
-
7. The plate was then allowed to solidify for 20 minutes.
8. Plate was then incubated for 48 hours @ 29 degrees C starting 1026 hrs. on 23/10/11.
-
9. Plate was removed from the incubator in order to check the status of the phage development at 1535 hrs. on 10/12/2023.
10. 23/10/13 Plates removed from incubator and placed in refrigerator for examination on Monday 23/10/16.
Results:
Results were positive. Plaques were produced on plates 10-1 and 10-2.
Conclusions and Next Steps:
Sample was positive for phages effecting M. foliorum bacteria in sufficient numbers to go to the next step. Begin amplification steps.
Title: Collecting Plate Lysates
Date: 10/16/23 Sample: Adopted 10-1 Plaque 4C Plate: 10-0
Purpose: To create a low concentration liquid sample of phage.
Notes:
- Using the 10^-2 plate we flooded it with 8 mL of phage buffer and allowed it to sit for three hours.
- After sitting, the plate was tilted and 5 mL of the lysate was drawn into a syringe.
- We then attached the syringe to a .22 micrometer filter and the fluid was pushed into a 15 mL conical tube.
- The tube was then labeled and ready for use.
We were able to collect 3 mL of our low volume lysate.
Results:
Collected Lysate from Adopted Plaque 4C
Conclusions and Next Steps:
9/29/2021: The lysate will now be used to make a full plate titer, and determine at what dilution will a webbed plate be produced, to then make a high titer lysate.
Title: Full Plate Titer
Date: 10/04/2021 Redo: No Sample: S9 Plaque A Plate: 10^-2
Purpose: To determine the concentration of phage in the lysate. So that, we can then determine what dilution will yield a webbed plate.
Notes:
- Using the low volume lysate collected we performed six ten fold dilutions. First, we added 90 microliters of phage buffer to each tube and then added ten microliters of our lysate to the 10^-1 tube and vortexed. Then, we took 10 microliters from the10^-1 tube and added it to the 10^-2. This was repeated to the 10^-6.
- We then did the plaque assay for each dilution. We added ten microliters of each tube into their respective 250 microliters of host bacteria and allowed ten minutes to set.
- We then added 3 mL of molten agar to each mixture and quickly aspirated it back and dispensed it onto agar plates avoiding bubbles.
- The plates were allowed to solidify for 20 mins and then incubated for 48 hours.
Results:
The full plate titer failed no plaques formed though bacterial growth was excellent
Conclusions and Next Steps:
10/18/2023: Since no plaques formed a new plate will be flooded to obtain a new lysate.
DNA Extraction
Title: DNA Extraction
Date: ##/###/##
Redo: Yes/No
Sample: #
Purpose: ###.
Notes:
Results:
###.
Conclusions and Next Steps:
###.
Title: DNA Extraction
Date: ##/###/##
Redo: Yes/No
Sample: #
Purpose: ###.
Notes:
Results:
###.
Conclusions and Next Steps:
###.
Characterization
Title: Setting Up Restriction Enzyme Digests
Date: ##/###/##
Redo: Yes/No
Sample: #
Purpose: ###.
Notes:
Results:
###.
Conclusions and Next Steps:
###.